 Author
Author  Correspondence author
Correspondence author
Field Crop, 2024, Vol. 7, No. 1
Received: 12 Dec., 2023 Accepted: 15 Jan., 2024 Published: 28 Jan., 2024
Genome-wide association study (GWAS) technology has become an important means to reveal the genetic basis of crop resistance. By analyzing a large number of genetic and phenotypic data, GWAS helps to identify key genes and genetic markers related to resistance. The aim of this study was to comprehensively analyze the latest progress of molecular genetic mechanism of barley (Hordeum vulgare L.) resistance by GWAS method, introduce the application basis of GWAS technology in plant science, and focus on its application cases in the study of barley resistance to abiotic stresses such as drought tolerance, salt tolerance and low temperature tolerance. This study will also explore current research challenges and look forward to the future direction of combining multi-omics data and advanced bioinformatics tools to more in-depth analysis of genetic mechanisms of barley resistance. This study provides a new perspective and strategy for improving barley resistance by molecular genetics, which is of great significance for the sustainable development of agriculture.
Barley (Hordeum vulgare L.), one of the pillars of global agriculture, plays an integral role, not only as a major food and feed source, but also in the beer and health food industries, with economic value and cultural significance across multiple sectors (Langridge, 2018). With the growth of the population and the diversification of consumption habits, the demand for barley continues to grow, and the growing environment of barley is facing unprecedented challenges. Extreme weather conditions brought about by global climate change, such as persistent droughts, frequent floods, salinized soils and abrupt temperature fluctuations, seriously threaten the growing cycle and yield of barley, thereby affecting global food security and the stability of agricultural economies.
In addressing these challenges, the application of modern genetics and molecular biology techniques has provided new directions for the improvement of barley. According to the study of Uffelmann et al. (2021), genome-wide association analysis (GWAS), as a powerful genetic analysis tool, has shown its unique advantages in the study of resistance to a variety of crops. By analyzing the relationship between genetic variation and phenotype, GWAS can identify genes or gene regions associated with specific traits without prior knowledge, and the application of this method has greatly promoted researchers' understanding of crop genetic diversity and accelerated the process of agricultural breeding.
A new breakthrough has been made in the study of barley resistance through GWAS technology. By analyzing barley samples from different environmental conditions, the researchers were able to identify a series of candidate genes associated with abiotic stresses such as drought tolerance, salt tolerance, and low temperature tolerance, the discovery of which not only enriched the understanding of barley stress response mechanisms, but also provided valuable resources for future molecular breeding (Gyawali et al., 2018). For example, through the functional verification of these resistance genes and the development of molecular markers, it is possible to achieve precise improvement of barley varieties for specific environmental stresses.
The GWAS study also revealed the complexity of barley resistance. Many reverse resistance traits have been found to be jointly regulated by multiple genes, which play roles in different physiological pathways, such as signal transduction, osmoregulation, antioxidant defense and hormone metabolism (Gyawali et al., 2018). This complex genetic network requires researchers to adopt a more systematic approach in future studies. For example, GWAS and other omics data were integrated to fully analyze the molecular mechanism of barley resistance.
Using GWAS to study the molecular genetic mechanism of barley resistance not only deepens the understanding of plant stress response mechanism, but also provides a new strategy for coping with global climate change and ensuring food security. With the advancement of genome sequencing technology and the development of bioinformatics tools, it is expected that more unknown resistance genes will be revealed in the future, providing a solid scientific foundation for the sustainable production of barley and other crops.
1 Overview of GWAS Technology
1.1 The fundamentals of GWAS technology
Genome-wide association analysis (GWAS) techniques have become a central tool in modern genetics and genomics research, especially in revealing the genetic basis behind complex traits. The basic principle of GWAS is to analyze the association between genetic variants (especially single nucleotide polymorphisms, SNPs) and phenotypes through statistical methods in order to identify genes or genetic regions that influence specific traits. The advantage of this approach is its genome-wide analytical capability, which enables researchers to discover new relevant genetic markers without prior knowledge of the genetic control mechanisms of the target trait (Marees et al., 2018) (Figure 1).
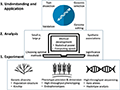 Figure 1 Challenges and opportunities in genome-wide association study (GWAS) (Cortes et al., 2021) |
The process of GWAS begins with precise measurements of the phenotypes and genotypes of a large number of individuals. Phenotypic data provide quantifiable information about studied traits, while genotypic data reveal the genetic variation of an individual across the whole genome, and use statistical methods to analyze these data to determine which genetic loci are significantly associated with phenotypic variation. These significantly correlated genetic loci are often considered candidate regions for influencing traits (Marees et al., 2018).
Although the application of GWAS in genetic research has achieved remarkable results, the technology also faces a series of challenges. GWAS requires large sample sizes to ensure adequate statistical power because genetic control of traits often involves multiple genes, each of which may have a relatively small effect. Genetic markers discovered by GWAS often require confirmation of their function through further bioinformatic analysis and experimental validation, a process that can be complex and time-consuming.
1.2 The development of GWAS technology
The development of genome-wide association analysis (GWAS) was an important milestone in the field of modern genetics and genomics, marking a key step in scientists' efforts to parse the genetic basis of complex traits. Since its first successful application to human genetic research in 2005, GWAS technology has experienced rapid development and wide application, which has greatly promoted the understanding of genetic mechanisms of polygenic diseases, crop traits, and other complex traits (Cortes et al., 2021) (Figure 2).
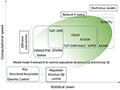 Figure 2 Genome-wide association study methods for improving computational speed and statistical power (Cortes et al., 2021) |
GWAS technology was born from several key technological advances: The development of high-throughput genotyping techniques made it possible to analyze genome-wide genetic variation in large numbers of samples at a reasonable cost. The creation of public databases and genetic information resources, such as the human genome project (HGP) and the HapMap Project, has provided GWAS with essential reference sequence and background genetic variation information. Advances in bioinformatics and statistical methods have provided tools for processing large-scale genetic data and complex statistical analyses .
Early GWAS research focused on human genetic diseases, successfully identifying multiple disease-related genetic markers, such as susceptibility genes for type 2 diabetes, coronary artery disease, and multiple cancers, and these findings not only revealed the genetic basis of disease, but also provided new ideas for disease prediction, prevention, and treatment. Later, the application of GWAS technology was gradually expanded to the agricultural field, and important agronomic traits of crops, such as yield, resistance and quality, were studied (Cortes et al., 2021). In crop research, GWAS has helped scientists identify key genes and genetic markers associated with traits, providing targets for crop molecular breeding and gene editing.
In recent years, with the further development of sequencing technology and the reduction of costs, GWAS has begun to evolve towards whole genome sequencing, which can capture genetic variation more comprehensively, improve the ability to find rare variants and resolve the genetic structure of complex traits, and integrate GWAS with other omics data, such as transcriptomic, proteomic and epigenetic data. It has become a new trend in current research. This multi-omics integration analysis is expected to further improve the analytical power of GWAS and reveal more complex genetic regulatory networks.
1.3 Application of GWAS in plant genetics
The application of genome-wide association analysis (GWAS) in the field of plant genetics has become an important means to reveal the genetic basis of plant traits. Since the first successful application of GWAS technology in the study of human genetic diseases, its application in plant science has expanded rapidly, and now covers a wide range of research fields from crop yield and quality improvement to stress tolerance. By analyzing correlations between genetic variation and phenotypic traits, this technique enables genome-wide identification of genes or genetic markers associated with important agronomic traits.
In terms of crop yield and quality improvement, GWAS has successfully identified many key genes that affect the yield and quality of food crops such as rice, wheat, maize, etc. These genes are involved in many aspects such as photosynthetic efficiency, nutrient absorption and utilization, grain size and component accumulation, and through the discovery and functional research of these key genes, scientists can carry out targeted molecular breeding to improve crop yield and quality (Liu and Yan, 2019).
In the study of stress tolerance, GWAS technology also shows its strong potential. With the intensification of global climate change, crops are facing increasing biological and abiotic stress, such as drought, salinity, low temperature, pests and diseases. By identifying genes associated with these stress responses, GWAS technology provides new ideas for revealing crop stress response mechanisms and developing stress-tolerant varieties (Lafarge et al., 2017). For example, using GWAS analysis, scientists identified key genes associated with drought response, salt stress tolerance, and cold resistance in multiple crops. GWAS technology has also been widely used in the study of plant growth and development, secondary metabolite synthesis, resistance to pests and diseases, etc. These studies not only enrich our understanding of plant physiological and ecological functions, but also provide scientific basis for crop genetic improvement and ecological environment protection.
2 Genetic Basis of Barley Resistance
2.1 Classification and genetic characteristics of inverse tolerance
In nature, plants are confronted with various biological and abiotic adversities, which seriously affect the growth, development and yield of plants. In this context, understanding plant resistance, especially for important crops such as barley, has become a key area of agricultural science research. Adverse tolerance can be divided into two main categories: abiotic and biological stress tolerance. Abiotic stresses include extreme climatic conditions such as drought, salinity, high or low temperatures, and heavy metal contamination in soil, while biological stresses involve the invasion of pathogens and pests (Singh et al., 2019). A plant's ability to respond and adapt to these adversities, known as resilience, is controlled by complex genetic mechanisms, often involving the interaction of multiple genes.
The genetic characteristics of reverse tolerance are characterized by polygenic control, variable gene effect sizes, environmental dependence, epistasis and phenotypic plasticity, which imply that reverse tolerance is a quantitative trait that is jointly influenced by multiple genes and environmental factors (Andersen et al., 2016). With the development of molecular biology techniques, especially the application of advanced methods such as genome-wide association analysis (GWAS), scientists are gradually revealing the genetic basis that controls resistance to reverse-resistance. These studies not only deepen researchers' understanding of the mechanisms by which plants survive and adapt to stress conditions, but also provide important molecular markers and candidate genes for the development of more resistant crop varieties.
2.2 Genetic markers associated with reverse tolerance in barley
In agricultural genetics and breeding research, identifying genetic markers associated with crop resistance is key to improving crop resilience and yield. Barley (Hordeum vulgare L.) is an important food and feed crop in the world, and its resistance to reverse-resistance has attracted extensive attention. Genetic markers related to resistance to adverse stress can not only help researchers understand the molecular mechanism of barley response to stress (Binott et al., 2017), but also provide a powerful tool for molecular-assisted breeding, especially in improving abiotic stress tolerance of barley such as drought, salinity and low temperature.
With the advancement of molecular biology techniques, especially the application of genome-wide association analysis (GWAS) and gene mapping techniques, researchers have successfully identified multiple genetic markers associated with resistance in the barley genome. These markers are usually located near key stress response genes or gene clusters, covering multiple levels such as signal transduction, gene expression regulation and metabolic pathway regulation. For example, some studies have found SNP markers related to drought tolerance in barley through GWAS analysis, which are located at or near known stress response genes. For example, genes in ABA (abscisic acid) signaling pathway, as well as some antioxidant oxidase genes, etc. (Tarawneh et al., 2020). In addition to abiotic stresses, the resistance of barley to pathogens is also an important aspect in the study of resistance to reverses. By locating genetic markers associated with resistance to specific diseases, the researchers were able to identify key genes that control barley disease resistance traits, such as resistance to rust, downy mildew and leaf spot.
2.3 Key resistance genes identified in previous studies
In the field of crop resistance research, previous studies have successfully identified several key genes that play a critical role in the response and adaptation mechanisms of plants in the face of abiotic and biological stresses, especially in important crops such as barley. For example, the DREB gene family, as transcription factors, plays a key role in regulating plant response to drought, low temperature and saline-alkali stress, and enhances plant adaptation by activating downstream stress response gene expression (Garrett et al., 2017). CBL/CIPK signaling system plays an important role in maintaining electrolyte balance and improving salt and drought tolerance in plant cells through its unique ion regulation mechanism.
Members of the LEA protein gene family play a protective role in plant resistance to drought and low temperature stress, and reduce the damage under adverse conditions by maintaining the water content of cells and the stability of biological macromolecules. In rice, the study of OsNHX1 gene reveals how plants respond to salt stress by regulating Na+/H+ exchange, which provides an important reference for the study of salt tolerance in other crops. The proline synthesis pathway involved in AtP5CS gene also shows its importance in plant resistance to drought and salt stress. By increasing proline synthesis, plants are able to enhance their osmoregulatory capacity and thus enhance stress tolerance.
3 Application of GWAS in the Study of Barley Resistance
3.1 A case study of barley drought tolerance tolerance by GWAS
In one study, scientists collected several barley germplasm resources and evaluated their phenotypic performance under drought conditions, including drought response traits such as leaf water potential, yield, and root structure. Illumina 9k single nucleotide polymorphism (SNP) chips were used for genome-wide SNP analysis of these barley germplasm.
Through GWAS analysis, the researchers identified multiple gene loci associated with drought tolerance in barley. These gene loci are located in different regions of the barley genome, some of which are associated with genes related to stress response such as root growth and ABA signaling pathways, and through further functional validation experiments, the researchers confirmed the importance of some candidate genes for drought tolerance in barley (Sallam et al., 2019).
The results of this study show that GWAS technology can help identify key genetic factors in barley drought tolerance and provide potential candidate genes for the future development of more drought-tolerant barley varieties, and the study also provides important clues for further understanding of barley drought response mechanisms. This case study shows that GWAS techniques have great potential for studying drought tolerance in barley and provide an important scientific basis for addressing environmental stress challenges such as drought. It is important to note that the GWAS findings require further validation and functional analysis to confirm the exact mechanism of action of candidate genes on drought tolerance in barley.
3.2 A case study of barley salt stress tolerance by GWAS
To carry out the GWAS study of salt tolerance in barley, the scientists collected a number of barley germplasm resources, which included barley species of different geographical origin and cultivation purposes. By conducting multiple phenotypic assessments under salt stress, such as growth indicators, biomass, ion concentration, etc., the researchers determined the level of tolerance of barley under salt stress.
A high density single nucleotide polymorphism (SNP) chip was used to analyze the whole genome SNP of these barley germplasm. Through GWAS analysis, the researchers found multiple gene loci related to barley salt stress tolerance, which were distributed in different regions of the barley genome (Fan et al., 2016), and through further functional verification experiments, the researchers identified some candidate genes. These genes are involved in stress response pathways related to salt stress response, ion balance and osmotic regulation.
This case study shows that GWAS techniques can help reveal the genetic basis of salt stress tolerance in barley and provide important clues for the future development of barley varieties with greater salt stress tolerance. The study also provides an important scientific basis for further understanding of barley's adaptation to salt stress, but the GWAS findings require further validation and functional analysis to confirm the exact mechanism of action of candidate genes on barley salt stress tolerance.
4 Challenges and Opportunities for GWAS Research on Barley Resistance
4.1 The effect of environmental variation on GWAS results
The influence of environmental variation on GWAS results is an important problem. The environmental conditions of barley growth may vary significantly due to geographical location, climate, soil texture and other factors, and such environmental variation may mask or distort genetic variation related to target traits. How to accurately control environmental factors to ensure the reliability and consistency of GWAS results is an urgent problem to be solved.
Another challenge is the complexity of the barley genome. The barley genome is large and complex, containing a large number of genes and genetic elements, and has complex features such as genome duplication and polymorphism, which makes it more difficult to identify genes and variants related to resistance in GWAS. Higher resolution genetic markers and more refined analytical methods are needed (Abdellaoui et al., 2022).
Despite the many challenges, GWAS research on barley resistance still contains great opportunities. With the continuous advancement of sequencing technology and the widespread application of single nucleotide polymorphism (SNP) chips, researchers are able to explore the genetic diversity of the barley genome more comprehensively, thus providing GWAS with richer genetic variation data. With the development of bioinformatics and statistical methods, researchers have also been able to more accurately control the influence of environmental factors and develop more accurate models of GWAS analysis, thus improving the accuracy and reliability of GWAS studies.
4.2 Analysis of genetic regulatory networks for complex traits
Genome-wide association study (GWAS), as an important genetic analysis tool, faces both challenges and great opportunities in the study of barley resistance. As one of the most important food crops, barley resistance is a key factor affecting yield and quality, which is of great significance for coping with climate change and improving crop resistance. When using GWAS to study barley resistance, we are faced with many challenges.
The genetic regulatory network of complex traits is an important challenge. Barley resistance is a complex trait, which is regulated by multiple genes and influenced by interaction with the environment. This complexity makes it more difficult to identify genes and variants associated with resistance in GWAS and requires a deep understanding of the genetic regulatory network of barley to reveal the genetic basis of resistance (Xu et al., 2022).
GWAS research on barley resistance is not only a challenging task, but also a field full of opportunities. Through in-depth understanding of the genetic regulatory network of barley, combined with advanced sequencing technology and analysis methods, it is expected to reveal the genetic basis of barley resistance, and provide an important scientific basis for breeding more resistant barley varieties.
4.3 The prospect of using GWAS results to guide molecular breeding
The prospect of using GWAS results to guide molecular breeding has attracted much attention in the agricultural field, and this approach provides new directions and opportunities for precision breeding. GWAS is a powerful tool for genetic analysis. By analyzing associations between large-scale genotype data and phenotypic data, genotypes and genes associated with target traits can be identified. Applying GWAS results to molecular breeding can accelerate the breeding process, improve breeding efficiency, and promote the cultivation of new varieties (Riaz et al., 2021).
Using GWAS results to guide molecular breeding can accelerate the discovery and utilization of high-quality genes. Through GWAS analysis of large natural populations, genotypes and genes associated with target traits can be quickly and accurately identified (Riaz et al., 2021). These high-quality genes can be directly used for the optimization of traditional breeding programs, and can also be used as the basis of molecular marker-assisted selection, so as to accelerate the utilization and transfer of high-quality genes.
Using GWAS results to guide molecular breeding can also improve breeding accuracy and prediction ability. Traditional breeding methods are often affected by complex environmental interactions between genotype and phenotype, resulting in low breeding efficiency. However, GWAS results can provide breeders with more accurate and reliable genetic information, help them better understand the genetic basis of target traits, and accurately predict the phenotypic performance of hybrid offspring (Spindel et al., 2016), so as to achieve the accurate achievement of breeding goals.
Using GWAS results to guide molecular breeding can also promote the innovation and development of breeding methods. With the continuous progress of molecular breeding technology, more and more molecular markers and analytical methods have been applied in breeding practice, providing breeders with more choices and possibilities. GWAS results provide important basis and support for the application of these molecular breeding technologies, and provide new ideas and directions for the innovation and development of breeding methods.
5 Outlook
With agricultural production facing more and more climate change and environmental pressure, the importance of research and improvement of reverse-resistant crops has become increasingly prominent. As one of the most important grain crops, barley resistance has attracted much attention. Using GWAS to study the molecular genetic mechanism of barley resistance is a prospective method, whose potential value is not only to reveal the genetic basis of barley resistance, but also to provide an important scientific basis for future precision breeding. Looking ahead, the following aspects will become the key development directions of GWAS research on barley resistance.
The integrated application of high-throughput sequencing technology will become an important trend in GWAS research. With the continuous development of sequencing technology, researchers can more comprehensively explore the genetic diversity of barley genome, thus providing more abundant genetic variation data. Combining high-throughput sequencing technology with GWAS can speed up the identification of genetic variation related to barley resistance, and provide scientific basis for further analysis of its molecular genetic mechanism.
Functional genomics will play an important role in the study of inverse resistance. By studying the function and regulatory mechanism of genes, functional genomics can deeply understand the mode of action and interrelationship of genes related to barley reverse-tolerance. Combined with the results of GWAS, functional genomics can help researchers understand the contribution degree of specific genes to barley reverse-tolerance, thus providing an important reference for the formulation of molecular regulatory strategies.
The application prospect of precision breeding technology in barley improvement is worth looking forward to. Precision breeding technology, such as gene editing and molecular marker-assisted selection, can accurately improve key genes identified by GWAS, and quickly breed barley varieties with more resistance. The application of precision breeding technology is expected to accelerate the breeding process of barley resistant varieties, improve their ability to adapt to environmental changes and withstand adversity pressure.
Using GWAS to study the molecular genetic mechanism of barley resistance is not only a challenging task, but also a field full of hope and prospects. By integrating high-throughput sequencing, the role of functional genomics, and the promise of precision breeding, researchers hope to uncover the underlying mechanisms of barley resistance and provide scientific basis for breeding more resistant barley varieties to address growing environmental challenges and ensure food security and sustainable agricultural development.
Abdellaoui A., Dolan C.V., Verweij K.J.H., and Nivard M.G., 2022, Gene-environment correlations across geographic regions affect genome-wide association studies, Nature Genetics, 54: 1345-1354.
https://doi.org/10.1038/s41588-022-01158-0
PMid:35995948 PMCid:PMC9470533
Andersen E.J., Ali S., Reese R.N., Yen Y., Neupane S., and Nepal M.P., 2016, Diversity and evolution of disease resistance genes in barley (Hordeum vulgare L.), Evolutionary Bioinformatics, 12: 99-108.
https://doi.org/10.4137/EBO.S38085
PMid:27168720 PMCid:PMC4857794
Binott J.J., Owuoche J.O., and Bartels D., 2017, Physiological and molecular characterization of Kenyan barley (Hordeum vulgare L.) seedlings for salinity and drought tolerance, Euphytica, 213: 139.
https://doi.org/10.1007/s10681-017-1924-2
Cortes L.T., Zhang Z., and Yu J., 2021, Status and prospects of genome-wide association studies in plants, The Plant Genome, 14(1): e20077.
https://doi.org/10.1002/tpg2.20077
PMid:33442955
Fan Y., Zhou G., Shabala S., Chen Z.H., Cai S., Li C., and Zhou M., 2016, Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.), Front. Plant Sci., 7: 946.
https://doi.org/10.3389/fpls.2016.00946
Garrett K.A., Andersen K.F., Asche F., Bowden R.L., Forbes G.A., Kulakow P.A., and Zhou B., 2017, Resistance genes in global crop breeding networks, Phytopathology, 107(10): 1268-1278.
https://doi.org/10.1094/PHYTO-03-17-0082-FI
PMid:28742460
Gyawali S., Chao S., Vaish S.S., Singh S.P., Rehman S., Vishwakarma S.R., and Verma R.P.S., 2018, Genome wide association studies (GWAS) of spot blotch resistance at the seedling and the adult plant stages in a collection of spring barley, Mol. Breeding, 38: 62.
https://doi.org/10.1007/s11032-018-0815-0
Lafarge T., Bueno C., Frouin J., Jacquin L., Courtois B., and Ahmadi N., 2017, Genome-wide association analysis for heat tolerance at flowering detected a large set of genes involved in adaptation to thermal and other stresses, PLoS ONE, 12(2): e0171254.
https://doi.org/10.1371/journal.pone.0171254
PMid:28152098 PMCid:PMC5289576
Langridge P., 2018, Economic and academic importance of barley, In: Stein N., and Muehlbauer G.J. (eds.), The Barley Genome, Compendium of Plant Genomes, Springer, Cham., Berlin, Germany, pp.1-10.
https://doi.org/10.1007/978-3-319-92528-8_1
Liu H.J., and Yan J., 2019, Crop genome-wide association study: a harvest of biological relevance, The Plant Journal, 97(1): 8-18.
https://doi.org/10.1111/tpj.14139
PMid:30368955
Marees A.T., de Kluiver H., Stringer S., Vorspan F., Curis E., Marie-Claire C., and Derks E.M., 2018, A tutorial on conducting genome-wide association studies: Quality control and statistical analysis, Psychiatric Research, 27(2): e1608.
https://doi.org/10.1002/mpr.1608
PMid:29484742 PMCid:PMC6001694
Riaz A., Kanwal F., Börner A., Pillen K., Dai F., and Alqudah A.M., 2021, Advances in genomics-based breeding of barley: Molecular tools and genomic databases, Agronomy, 11(5): 894.
https://doi.org/10.3390/agronomy11050894
Sallam A., Alqudah A.M., Dawood M.F.A., Baenziger P.S., and Börner A., 2019, Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research, Int. J. Mol. Sci., 20(13): 3137.
https://doi.org/10.3390/ijms20133137
PMid:31252573 PMCid:PMC6651786
Singh B., Mehta S., Aggarwal S.K., Tiwari M., Bhuyan S.I., Bhatia S., and Islam M.A., 2019, Barley, disease resistance, and molecular breeding approaches, In: Wani S.H. (ed.), Disease resistance in crop plants, Springer, Cham., Beilin, Germany, pp.261-299.
https://doi.org/10.1007/978-3-030-20728-1_11
Spindel J.E., Begum H., Akdemir D., Collard B., Redoña E., Jannink J.L., and McCouch S., 2016, Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement, Heredity, 116: 395-408
https://doi.org/10.1038/hdy.2015.113
PMid:26860200 PMCid:PMC4806696
Tarawneh R.A., Alqudah A.M., Nagel M., and Börner A., 2020, Genome-wide association mapping reveals putative candidate genes for drought tolerance in barley, Environmental and Experimental Botany, 180: 104237.
https://doi.org/10.1016/j.envexpbot.2020.104237
Uffelmann E., Huang Q.Q., Munung N.S., de Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., and Posthuma D., 2021, Genome-wide association studies, Nature Reviews Methods Primers, 1: 59.
https://doi.org/10.1038/s43586-021-00056-9
Xu Q., Huang S., Guo G., Yang C., Wang M., Zeng X., and Wang Y., 2022, Inferring regulatory element landscapes and gene regulatory networks from integrated analysis in eight hulless barley varieties under abiotic stress, BMC Genomics, 23: 843.
https://doi.org/10.1186/s12864-022-09070-x
PMid:36539685 PMCid:PMC9769044
. HTML
Associated material
. Readers' comments
Other articles by authors
. Heming Wei
Related articles
. High-throughput sequencing
. Epigenetics
. Disease research
. DNA methylation
. Personalized medicine
Tools
. Post a comment


