Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2010, Vol. 1, No. 1 doi: 10.5376/rgg.2010.01.0001
Received: 10 Aug., 2010 Accepted: 02 Sep., 2010 Published: 08 Oct., 2010
He et al., 2009, Characterization and Mapping of Rice Mutants, des2 and des5, Genomics and Applied Biology, 28(1): 1-9
Identification of key regulator genes in the control of spikelet development in panicle, one of the main agronomic traits to determine rice yield, is important to investigate the underlying molecular mechanism and provide the molecular basis for analysis of the agronomic traits. In this study, we have identified a group of mutants with conspicuous reduction of spikelet number, which are designated decreased spikelets (des). Two of the des mutants, des2 and des5, were analyzed in detail. We showed that a single recessive locus was responsible for des2 phenotype, which is located on the long arm of chromosome 6. The des2 locus was cloned and it was found that des2 is a new allele of moc1. Positional cloning and sequence analysis showed that there is a point mutation in the HLH domain-encoding region of LAX gene in des5, indicating that des5 is a new allele of lax. Our data indicate that spikelet development is regulated by the same pathway in the control of development of axillary branches in rice.
Many agronomic traits of crops, such as plant height, tillering, panicle morphology and grain weight, are controlled both genetically and environmentally. Rice (Oryza sativa L.), one of the most important staple foods in the world, has been extensively studied as model plant and significant progress has been made to investigate the key loci in the control of main agronomic traits (Monna et al., 2002; Sasaki et al., 2002a; Spielmeyer et al., 2002; Hedden, 2003; Song et al., 2007; Li et al., 2003; Komatsu et al., 2003). A great number of regulatory genes and QTL were cloned by forward genetics (from phenotype to gene), which was greatly facilitated by the availability of whole genome sequence in rice (Feng et al., 2002; Goff et al., 2002; Kikuchi et al., 2003; International Rice Genome Sequencing Project, 2005; The Rice Chromosome 10 Sequencing Consortium, 2003; Sasaki et al., 2002b; Shimamoto and Kyozuka, 2002; Yu et al., 2002; Zhang et al., 2004). Three groups independently identified “green revolution gene”, SEMIDWARF1 (SD-1) which encodes a GA 20-oxidase (GA20ox-2) and catalyzes a key step in GA biosynthesis process (Monna et al., 2002; Sasaki et al., 2002a; Spielmeyer et al., 2002; Hedden, 2003). The dwarfism (reduced plant height) conferred by sd-1 mutant is a desirable trait that enhances the ability of plant's resistance to lodging (or falling-over), thus reducing yield losses. Apart from genetic studies on plant height, a great number of QTL responsible for grain weight were also identified. A major QTL, named GW2 which encodes a RING-finger-containing protein, plays a significant role in the control of grain weight (Song et al., 2007). Some rice varieties, such as WY3 and Oochikara, with reduction or loss of function of GW2, results in improved grain weight.
The genetic regulation of tillering or branching and panicle morphology has been a main focus for researchers. monoculm 1 (moc1) mutant has a single main culm with few tiller, indicating an important locus in the control of tillering in rice. Positional cloning revealed that MOC1 encodes a transcription factor belonging to the GRAS family. It is demonstrated that MOC1 may function as a master regulator to determine tillering by modulating the expression of the other two transcription factors, OSH1 and OsTB1 (Li et al., 2003). It has been reported that there were multiple genetic components in the control of branching during rice development. lax panicle (lax) possesses the phenotype of reduced number of axillary branches (Komatsu et al., 2001; 2003). It has been shown that LAX encodes a bHLH transcription factor. Another mutant, small panicle (spa) displays an abnormal phenotype with a smaller panicle but with normal tiller number. However, when the spa was introduced into lax-1 background, a weak allele of lax, the double mutant exhibited extremely deficiency in tiller number and had a single main culm bearing a barren panicle. It was proposed that all axillary branch development should be controlled by a single genetic mechanism (Komatsu et al., 2003). In aberrant panicle organization 1 (apo1), the precocious termination of the main-axis meristem and the precocious conversion of primary branch meristem into spikelet meristem results in short rachis and reduces the number of primary branches (PB) and spikelets significantly (Ikeda et al., 2005; 2007). APO1 has been cloned and found to encode an F-box protein, indicating that ubiquitin-proteasome pathway may be involved in regulating branch development (Ikeda et al., 2005; 2007). It has been well established that SCF complex constituted by Skp1p, Cullin and F-box protein can ubiquitinate and target specific substrate to 26S proteasome for degradation (Deshaies, 1999; Smalle and Vierstra, 2004; Moon et al., 2004; Gagne et al., 2002; Pickart, 2001; Risseeuw et al., 2003; Cardozo and Pogano, 2004; Zheng et al., 2002). Thus, posttranslational modification of some key factors or the interaction at protein level should play an important role during axillary branching. Recently, a pair of enzymes regulating cytokinin level but with opposite function were identified to control panicle morphology (Ashikari et al., 2005; Kurakawa et al., 2007), indicating phytohormones is implicated in axillary branch development.
Although quite a few regulatory genes controlling axillary branch development have been identified, the underlying genetic network just emerges and more key regulators are expected to be discovered. In this study, we identified a group of mutants, decreased spikelets (des), which have conspicuous effect to reduce the spikelet number. Among these des loci, DES2 was cloned by map-based cloning approach and analyzed in detail. It was found that des2 is allelic to MOC1. In des2, a point mutation occurred in the 3' end of coding region in MOC1, leading to a premature translation stop and giving rise to a deletion of the last 5 amino acid residues in C terminus of MOC1. des5 was identified as a new allele of lax, and carries a point mutation at the HLH domain. Furthermore, it was found that des2 and des5 could interact with each other, abolishing all axillary branch development. Out data indicate that MOC1 and LAX should interplay with each other in the control of development of axillary branches in rice.
1 Materials and methods
1.1 Plant materials
Four des mutants, des1, des2, des3 and des4, were identified from an M2 mutagenized japonica rice cultivar Zhonghua 11 (ZH11) by γ-ray irradiation and des5 from another one by EMS mutagenesis. The mapping population was derived from F2 segregation population of the cross between des2 and the indica rice cultivar Longtefu.
1.2 DNA extraction
Total DNA was extracted from young rice leaves according to the CTAB method (Murray and Thompson, 1980), with some modifications. About 0.1 g rice leaves were ground into powder in liquid N2. Add 400 μL extraction buffer and incubate in 65℃ water bath for 30~60 min with occasional gentle mixing. Add 400 μL chloroform and shake followed by centrifugation at 12 000 rpm for 5 minutes. Transfer the supernatant into a new centrifugal tube and add 0.8~1 volume of cold isopropanol with mixing. Place it at -20℃ for 15 min followed by centrifugation at 12 000 rpm for 5 minutes. Pour off the supernatant and wash the pellet with 500 μL 70% ethanol once. After drying in the air, add 100 μL TE or sterile water to dissolve DNA.
1.3 Marker analysis by polyacrylamide gel electrophoresis (PAGE)
The PCR reaction system (20 μL) consists of 2 μL 10×buffer (supplied with Taq polymerase), 0.2 mmol/L of dNTP, 5% dimethyl sulfoxide (DMSO), 20 ng of DNA, 0.1 μmol/L of primers and 1 U of Taq polymerase. The PCR program for SSR and dCAPS markers is as follows: 94℃ for 3 min, followed by 35 cycles of 94℃ for 30 s, annealing (temperature adjusted with different primers) for 30 s, 72℃ for 30 s, and finally 72℃ for 10 min. The primer sequences of these SSR and STS markers were downloaded from Gremene (http://www.gramene.org) and RGP (http://rgp.dna.affrc.go.jp/E/index.html) respectively. The restriction endonuclease digestion for dCAPS markers was conducted in a 20 μL reaction mixture containing 2 μL 10×buffer (supplied with enzyme), 0.2 μL 100×BSA when necessary, 0.1~0.5 μg of PCR products and 15~20 U of restriction enzyme at the optimal temperature for 2~3 hours. The polymorphisms of these markers were analyzed on 6% polyacrylamide gel stained with 0.1% silver nitrate.
1.4 New molecular markers in des2 region
SSR markers listed in Gramene database are normally tested and used in this study. In the cases when new markers were needed, they were developed by either optimizing the ones from the database or comparing the public available japonica and indica sequences (Feng et al., 2002; Goff et al., 2002; Kikuchi et al., 2003; International Rice Genome Sequencing Project, 2005; Sasaki et al., 2002b; The Rice Chromosome 10 Sequencing Consortium, 2003; Shimamoto and Kyozuka, 2002; Yu et al., 2002; Zhang et al., 2004) so as to design new markers including SSR and dCAPS in potential polymorphic sites (Table 1).
|
Table 1 Newly-developed molecular markers
|
1.5 Construction of linkage map
Linkage map was constructed by using MapDraw V2.1 (Liu and Meng, 2003).
2 Results
2.1 Phenotype of des mutants
To investigate the key regulators in the control of spikelet development, we screened rice mutants with the phenotype of significant reduction of spikelet number, which were designated decreased spikelets (des). Five complementary groups of des mutants, des1, des2, des3, des4 and des5, were identified in the mutagenized M2 population of ZH11. In the des mutants, the reduction rate of spikelets was typically about 30%~40% reduction in comparison to the wild type (Figure 1 and Table 2). However, in des mutants the development of tillers, as well as primary branches (PB) and secondary branches (SB), were also affected to various extents (Figure 1 and Table 2). Among these des mutants, des2 and des5 were characterized in detail in this study.
|
Figure 1 The effects of des2 and des5 on tillering and panicle
|
|
Table 2 Comparison of the traits of axillary branches among genotypes
|
2.2 Genetic analysis and primary mapping of des2
Genetic analysis was conducted to determine whether des2 is controlled by a single locus. In a cross between des2 and ZH11, the segregation ratio of wild type and mutants in the F2 population was in accordance with 3:1 (n=126, χ2=0.01<χ20.05,1=3.84). This indicates that a single recessive locus is responsible for des2 (Table 3).
|
Table 3 Segregation analysis among F2 population
|
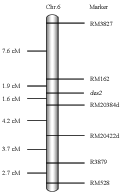
A mapping population was constructed by crossing des2 with an indica variety, Longtefu. 147 individuals with typical des2 phenotype in F2 population were selected for des2 mapping. Primarily, 15 mutant individuals were randomly selected to comprise a DNA pool, with which 92 SSR markers evenly distributed in 12 chromosomes were subjected to tests for linkage with des2 locus. As a result, two SSR markers RM3827 and RM528 on chromosome 6 were found to be linked with des2, which was further confirmed by testing some F2 individuals with these two markers. Then the candidate region was further framed between RM162 and a STS marker R3879. Furthermore, new molecular markers including SSR and dCAPS were developed by analyzing the japonica and indica genome sequences and their polymorphisms between ZH11 and Longtefu were analyzed in PAGE (Table 1). By developing the new markers, des2 was confined between RM162 and RM20384d (short for RM20384-derived. See Materials and Methods) with estimated genetic distances of 1.9 cM and 1.6 cM respectively (Figure 2).
|
Figure 2 Genetic map of the region spanning des2 locus on chromosome 6
|
2.3 Fine mapping of des2
A larger mapping population was constructed for fine mapping of des2, from which 500 mutant plants were subjected to the analysis. Then the locus was delimited to a 28 kb region between two markers OsdCAPS2 and OsdCAPS3, and the des2 locus was found to co-segregate with a SSR marker RM20378d since there was no recombinant among the 500 mutant plants. A PAC contig was found to span the des2 locus (Figure 3). Within the delimited region, only one gene encoding a hypothetic protein and the previously identified MOC1 gene were found, according to the annotation in NCBI (http://www.ncbi.nlm.nih.gov). No mutation was detected in the unknown gene by sequencing, however, there was a point mutation was found in the 3' end of MOC1 coding region, leading to a premature translation stop and giving rise to a truncated MOC1 protein with a deletion of the last 5 amino acid residues in C terminus (Figure 4A). Therefore, des2 phenotype should be caused by the point mutation in MOC1 gene.
|
Figure 3 The fine map encompassing des2 locus on chromosome 6 and PAC contig spanning des2 locus
|
2.4 des5 being allelic to LAX
Apart from the reduction of spikelet number, defects on panicle development with less SB and SP are also observed in des5 (Table 2). Genetic analysis showed that a single locus should be mutated and lead to the defects in des5 in a recessive manner (n=140, χ2=0.35<χ20.05,1=3.84) (Table 3). Genetic analysis of des5 locus was conducted and positional cloning delimited des5 to a region between markers RSL0081 and RM3640 on the long arm of chromosome 1 (data not shown). Sequence analysis indicated that LAX is located inside the region, which has been shown to play a key role in the control of branch formation during panicle development in rice (Komatsu et al., 2001). Later it was found that in des5 there is a point mutation in the coding region of LAX gene, giving rise to a substitution of Phe-63 for Leu-63 in its HLH domain (Figure 4B). It is expected that the mutated lax should not have a proper function and be consistent with the abnormal panicle development in des5. Therefore, we conclude that des5 should be a new allele of lax mutant.
|
Figure 4 The mutation sites in des2 and des5
|
3 Discussion
In this study, we identified a few des mutants, which mainly affect the spikelet development but with relatively minor effects on development of tiller and branch in panicle. Two loci, des2 and des5, were cloned and the others are subjected to analysis in detail at the moment. It is found that des2 and des5 are allelic to the regulatory genes, MOC1 and LAX, respectively.
The des5, displays very similar or consistent phenotype as the one of lax1 (Komatsu et al., 2001). It has been found that LAX encodes a bHLH transcription factor and regulates panicle development by monitoring the initiation of axillary primordium (Komatsu et al., 2001; 2003). In des5, there is a point mutation at the HLH domain of LAX, which presumably has a key function for protein-protein interaction and forms either homodimer or heterodimer (Heim et al., 2003). For example, SPCH, MUTE and FAMA are three key bHLH-type transcription factors regulating stomatal development (MacAlister et al., 2007; Pillitteri et al., 2007; Ito and Bergmann, 2006). It has been shown that their individual function requires physical interaction with two other bHLH-type transcription factors SCREAM1 and SCREAM2; moreover, yeast two-hybrid demonstrates that MUTE itself can strongly form homodimer (Kanaoka et al., 2008). Consistently, our data suggest that the Leu-63 at the HLH domain should be essential for the protein function of LAX.
MOC1 has been previously reported as a key factor in the control of tiller development (Li et al., 2003). However, in the case of des2, there are significant differences between the phenotypes of des2 and the previously reported moc1. The phenotype of a single culm without tiller in moc1 is contrasted to the relatively normal tillering but conspicuous effect on spikelet number in des2. These could be due to the different genetic background, or alternatively could be explained by the allelic difference. The lesion of MOC1 protein in des2 mutant is subtle in comparison with the one in moc1. In des2, the last5 amino acid residues in the C terminus of MOC1 is missing, whereas the last 125 residues were deleted in moc1 mutant with an extra 1.9 kb insertion of a retrotransposon (Li et al., 2003). Therefore, des2 can be considered as a weak allele of moc1. Nevertheless, the serine homotrimeric stretch is comprised in the last 5 residues of MOC1 which is lost in des2. It is likely that these serine residues are extra potential phosphorylation sites besides the conserved tyrosine (Li et al., 2003). Our data indicate the importance of the C terminus for the protein function of MOC1. In des2 mutant, the conspicuous phenotype of substantial reduction of the spikelet number in panicle, but relatively minor effect on the number of tiller and branches in panicle, suggest that the action of MOC1 could be differentiated depending on the developmental stages. During rice development, the axillary meristem acquires distinct identities in vegetative and reproductive development. It is possible that different axillary meristem development requires differential levels of MOC1 activity, or different domains of MOC1 protein play different roles, which should be essential to interact with other key regulators in the control of formation of tiller, PB, SB and spikelet, respectively.
While the strong phenotype of moc1 could be an obstacle to investigate the genetic interaction between MOC1 and other key regulators, the weak phenotype of des2 was explored in this study. Our preliminary data indicate that different combinations of different des double mutants display enhanced phenotype (He et al., unpublished data), suggesting that the interplay between different DES genes should be a common theme. The synergistic effect of different double des mutants could shed some light on the potential mode of DES function. It is possible that different DESs could be involved in several distinct pathways and act independently to control axillary branch development. Alternatively, MOC1 and LAX should function as partners in a common pathway. However, these two possibilities are not exclusive. In phytochrome A signaling pathway, two positive regulators LAF1 and HFR1 have both independent and interdependent functions with respect to the inhibition of hypocotyl elongation by far-red light (Jang et al., 2007). It was shown that COP1-targeted LAF1 and HFR1 protein degradation was inhibited by their physical association (Jang et al., 2007). There have been several reports indicating the regulation of shoot branching at protein level and when an F-box protein, APO1 is mutated, the development of PB and spikelet is affected (Ikeda et al., 2005; 2007). Thus, the posttranslational modification of some key factors or interaction at protein level might play an important role in regulating the formation of branching and spikelet in panicle. MOC1 belongs to the GRAS family and consistently is found to be localized in nucleus. LAX is a bHLH transcription factor and its function could be dependent on its heterodimeric status. In the future work, it is feasible to test whether there is direct protein-protein interaction between LAX and MOC1. It has been proposed that development of axillary branches is controlled by a single genetic mechanism (Komatsu et al., 2003). Therefore, our data is consistent with this notion since most of the spikelets are initiated as the axillary primordium in the periphery zone in the apical meristems of PB and SB. Our data indicates that spikelet development is regulated by the same pathway in the control of development of axillary branches in rice. With more des loci being identified and more DES genes cloned, the complexity of the genetic network in the control of spikelet development should be investigated in both genetic and protein levels.
Acknowledgements
This study was supported by the National High Technology Research and Development Program of China (2006AA10A102) and the National Key Project on Basic Sciences (2005CB120800)
References
Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., and Matsuoka M., 2005, Cytokinin oxidase regulates rice grain production, Science, 309(5735): 741-745
doi:10.1126/science.1113373
Cardozo T., and Pagano M., 2004, The SCF ubiquitin ligase: insights into a molecular machine, Nat. Rev. Mol. Cell Biol., 5(9): 739-751 doi:10.1038/nrm1471
Deshaies R.J., 1999, SCF and Cullin/Ring H2-based ubiquitin ligases, Annu. Rev. Cell Dev. Biol., 15: 435-467
doi:10.1146/annurev.cellbio.15.1.435
Feng Q., Zhang Y., Hao P., Wang S., Fu G., Huang Y., Li Y., Zhu J., Liu Y., Hu X., Jia P., Zhang Y., Zhao Q., Ying K., Yu S., Tang Y., Weng Q., Zhang L., Lu Y., Mu J., Lu Y., Zhang L.S., Yu Z., Fan D., Liu X., Lu T., Li C., Wu Y., Sun T., Lei H., Li T., Hu H., Guan J., Wu M., Zhang R., Zhou B., Chen Z., Chen L., Jin Z., Wang R., Yin H., Cai Z., Ren S., Lv G., Gu W., Zhu G., Tu Y., Jia J., Zhang Y., Chen J., Kang H., Chen X., Shao C., Sun Y., Hu Q., Zhang X., Zhang W., Wang L., Ding C., Sheng H., Gu J., Chen S., Ni L., Zhu F., Chen W., Lan L., Lai Y., Cheng Z., Gu M., Jiang J., Li J., Hong G., Xue Y., and Han B., 2002, Sequence and analysis of rice chromosome 4, Nature, 420(6913): 316-320 doi:10.1038/nature01183
Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., and Vierstra R.D., 2002, The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis, Proc. Natl. Acad. Sci., USA, 99(17): 11519-11524 doi:10.1073/pnas.162339999
Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Hadley D., Hutchison D., Martin C., Katagiri F., Lange B.M., Moughamer T., Xia Y., Budworth P., Zhong J., Miguel T., Paszkowski U., Zhang S., Colbert M., Sun W.L., Chen L., Cooper B., Park S., Wood T.C., Mao L., Quail P., Wing R., Dean R., Yu Y., Zharkikh A., Shen R., Sahasrabudhe S., Thomas A., Cannings R., Gutin A., Pruss D., Reid J., Tavtigian S., Mitchell J., Eldredge G., Scholl T., Miller R.M., Bhatnagar S., Adey N., Rubano T., Tusneem N., Robinson R., Feldhaus J., Macalma T., Oliphant A., and Briggs S., 2002, A draft sequence of the rice genome (Oryza sativa L. ssp. japonica), Science, 296(5565): 92-100 doi:10.1126/science.1068275
Hedden P., 2003, The genes of the green revolution, TRENDS in Genetics, 19(1): 5-9 doi:10.1016/S0168-9525(02)00009-4
Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., and Bailey P.C., 2003, The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity, Mol. Biol. Evol., 20(5): 735-747 doi:10.1093/molbev/msg088
Ikeda K., Ito M., Nagasawa N., Kyozuka J., and Nagato Y., 2007, Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate, Plant J., 51(6): 1030-1040 doi:10.1111/j.1365-313X.2007.03200.x
Ikeda K., Nagasawa N., and Nagato Y., 2005, ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice, Developmental Biology, 282(2): 349-360 doi:10.1016/j.ydbio.2005.03.016
International Rice Genome Sequencing Project, 2005, The map-based sequence of the rice genome, Nature, 436(7052): 793-800 doi:10.1038/nature03895
Ito K.O., and Bergmann D.C., 2006, Arabidopsis FAMA Controls the final proliferation/differentiation switch during stomatal development, The Plant Cell, 18(10): 2493-2505 doi:10.1105/tpc.106.046136
Jang I.C., Yang S.W., Yang J.Y., and Chua N.H., 2007, Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling, Genes Dev., 21(16): 2100-2111 doi:10.1101/gad.1568207
Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., and Torii K.U., 2008, SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation, The Plant Cell, 20(7): 1775-1785 doi:10.1105/tpc.108.060848
Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Hotta I., Kojima K., Namiki T., Ohneda E., Yahagi W., Suzuki K., Li C.J., Ohtsuki K., Shishiki T., Otomo Y., Murakami K., Iida Y., Sugano S., Fujimura T., Suzuki Y., Tsunoda Y., Kurosaki T., Kodama T., Masuda H., Kobayashi M., Xie Q., Lu M., Narikawa R., Sugiyama A., Mizuno K., Yokomizo S., Niikura J., Ikeda R., Ishibiki J., Kawamata M., Yoshimura A., Miura J., Kusumegi T., Oka M., Ryu R., Ueda M., Matsubara K., Kawai J., Carninci P., Adachi J., Aizawa K., Arakawa T., Fukuda S., Hara A., Hashizume W., Hayatsu N., Imotani K., Ishii Y., Itoh M., Kagawa I., Kondo S., Konno H., Miyazaki A., Osato N., Ota Y., Saito R., Sasaki D., Sato K., Shibata K., Shinagawa A., Shiraki T., Yoshino M., Hayashizaki Y., and Yasunishi A., 2003, Collection, mapping and annotation of over 28 000 cDNA clones from japonica rice, Science, 301(5631): 376-379 doi:10.1126/science.1081288
Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., and Kyozuka J., 2003, LAX and SPA: major regulators of shoot branching in rice, Proc. Natl. Acad. Sci., USA, 100(20): 11765-11770 doi:10.1073/pnas.1932414100
Komatsu M., Maekawa M., Shimamoto K., and Kyozuka J., 2001, The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development, Developmental Biology, 231(2): 364-373 doi:10.1006/dbio.2000.9988
Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., and Kyozuka J., 2007, Direct control of shoot meristem activity by a cytokinin-activating enzyme, Nature, 445(7128): 652-655 doi:10.1038/nature05504
Li X.Y., Qian Q., Fu Z.M., Wang Y.H., Xiong G.S., Zeng D.L., Wang X.Q., Liu X.F., Teng S., Hiroshi F., Yuan M., Luo D., Han B., and Li J.Y., 2003, Control of tillering in rice, Nature, 422(6932): 618-621 doi:10.1038/nature01518
Liu R.H., and Meng J.L., 2003, MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data, Heraditas, 25(3): 317-321
MacAlister C.A., Ito K.O., and Bergmann D.C., 2007, Transcription factor control of asymmetric cell divisions that establish the stomatal lineage, Nature, 445(7127): 537-540 doi:10.1038/nature05491
Monna L., Kitazawa N., Yoshino R., Suzuki J., Masuda H.,Maehara Y., Tanji M., Sato M., Nasu S., and Minobe Y., 2002, Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis, DNA Research, 9(1): 11-17 doi:10.1093/dnares/9.1.11
Moon J., Parry G., and Estelle M., 2004, The ubiquitin-proteasome pathway and plant development, The Plant Cell, 16(12): 3181-3195 doi:10.1105/tpc.104.161220
Murray M.G., and Thompson W.F., 1980, Rapid isolation of high molecular weight plant DNA, Nucleic Acids Research, 8(19): 4321-4325 doi:10.1093/nar/8.19.4321
Pickart C.M., 2001, Mechanisms underlying ubiquitination, Annu. Rev. Biochem, 70: 503-533 doi:10.1146/annurev.biochem.70.1.503
Pillitteri L.J., Sloan D.B., Bogenschutz N.L., and Torii K.U., 2007, Termination of asymmetric cell division and differentiation of stomata, Nature, 445(7127): 501-505 doi:10.1038/nature05467
Risseeuw E.P., Daskalchuk T.E., Banks T.W., Liu E., Cotelesage J., Hellmann H., Estelle M., Somers D.E., and Crosby W.L., 2003, Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis, Plant J, 34(6): 753-767 doi:10.1046/j.1365-313X.2003.01768.x
Sasaki A., Ashikari M., Tanaka M.U., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., Kitano H., and Matsuoka M., 2002a, Green revolution: a mutant gibberellin-synthesis gene in rice, Nature, 416(6882): 701-702 doi:10.1038/416701a
Sasaki T., Matsumoto T., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Antonio B.A., Kanamori H., Hosokawa S., Masukawa M., Arikawa K., Chiden Y., Hayashi M., Okamoto M., Ando T., Aoki H., Arita K., Hamada M., Harada C., Hijishita S., Honda M., Ichikawa Y., Idonuma A., Iijima M., Ikeda M., Ikeno M., Ito S., Ito T., Ito Y., Ito Y., Iwabuchi A., Kamiya K., Karasawa W., Katagiri S., Kikuta A., Kobayashi N., Kono I., Machita K., Maehara T., Mizuno H., Mizubayashi T., Mukai Y., Nagasaki H., Nakashima M., Nakama Y., Nakamichi Y., Nakamura M., Namiki N., Negishi M., Ohta I., Ono N., Saji S., Sakai K., Shibata M., Shimokawa T., Shomura A., Song J., Takazaki Y., Terasawa K., Tsuji K., Waki K., Yamagata H., Yamane H., Yoshiki S., Yoshihara R., Yukawa K., Zhong H., Iwama H., Endo T., Ito H., Hahn J.H., Kim H.I., Eun M.Y., Yano M., Jiang J., and Gojobori T., 2002b, The genome sequence and structure and rice chromosome 1, Nature, 420(6913): 312-316 doi:10.1038/nature01184
Shimamoto K., and Kyozuka J., 2002, Rice as a model for comparative genomics of plants, Annu. Rev. Plant Biol., 53: 399-419 doi:10.1146/annurev.arplant.53.092401.134447
Smalle J., and Vierstra R.D., 2004, The ubiquitin 26S proteasome proteolytic pathway, Annu. Rev. Plant Biol., 55: 555-590 doi:10.1146/annurev.arplant.55.031903.141801
Song X.J., Huang W., Shi M., Zhu M.Z., and Lin H.X., 2007, A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase, Nature Genetics, 39(5): 623-630 doi:10.1038/ng2014
Spielmeyer W., Ellis M.H., and Chandler P.M., 2002, Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene, Proc. Natl. Acad. Sci., USA, 99(13): 9043-9048 doi:10.1073/pnas.132266399
The Rice Chromosome 10 Sequencing Consortium, 2003, In-depth view of structure, activity, and evolution of rice chromosome 10, Science, 300(5625): 1566-1569
Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Cao M., Liu J., Sun J., Tang J., Chen Y., Huang X., Lin W., Ye C., Tong W., Cong L., Geng J., Han Y., Li L., Li W., Hu G., Huang X., Li W., Li J., Liu Z., Li L., Liu J., Qi Q., Liu J., Li L., Li T., Wang X., Lu H., Wu T., Zhu M., Ni P., Han H., Dong W., Ren X., Feng X., Cui P., Li X., Wang H., Xu X., Zhai W., Xu Z., Zhang J., He S., Zhang J., Xu J., Zhang K., Zheng X., Dong J., Zeng W., Tao L., Ye J., Tan J., Ren X., Chen X., He J., Liu D., Tian W., Tian C., Xia H., Bao Q., Li G., Gao H., Cao T., Wang J., Zhao W., Li P., Chen W., Wang X., Zhang Y., Hu J., Wang J., Liu S., Yang J., Zhang G., Xiong Y., Li Z., Mao L., Zhou C., Zhu Z., Chen R., Hao B., Zheng W., Chen S., Guo W., Li G., Liu S., Tao M., Wang J., Zhu L., Yuan L., and Yang H., 2002, A draft sequence of the rice genome (Oryza sativa L. ssp. indica), Science, 296(5565): 79-92 doi:10.1126/science.1068037
Zhang Y., Huang Y., Zhang L., Li Y., Lu T., Lu Y., Feng Q., Zhao Q., Cheng Z., Xue Y., Wing R.A., and Han B., 2004, Structural features of the rice chromosome 4 centromere, Nucleic Acids Res., 32(6): 2023-2030 doi:10.1093/nar/gkh521
Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., Conaway R.C., Conaway J.W., Harper J.W., and Pavletich N.P., 2002, Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex, Nature, 416(6882): 703-709 doi:10.1038/416703a
. PDF(344KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Shengbo He
. Wei He
. Hongqi Chen
. Xudong Zhu
. Jun Yang
. Xiaohe Hu
. Da Luo
Related articles
. Rice mutants
. des2
. des5
. Characterization
. Mapping
Tools
. Email to a friend
. Post a comment
.png)