Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2012, Vol. 3, No. 7 doi: 10.5376/rgg.2012.03.0007
Received: 21 May, 2012 Accepted: 27 Jun., 2012 Published: 29 Jun., 2012
Hu et al., 2012, Genetics and Molecular Breeding for Salt-Tolerance in Rice, Vol.3, No.7 39-38 (doi: 10.5376/rgg.2012.03.0007)
Salt stress is one of the main environmental constraints for the losses of rice yield. In this paper, we introduced the mechanisms of rice salt-tolerance by the following three aspects: the integrity of membrane systems, ionic compartmentation, and osmotic adjustment. We also briefly presented the three methods for identifying rice salt-tolerance, which specifically refer to biological and agronomic salt resistance, as well as the response of in vitro cells to salt stress. Then we summarized the progresses of mining salt-resistance rice germplasm resources, mapping the QTLs conferring salt-tolerance, cloning slat-tolerant genes of importance and breeding salt-tolerant rice varieties. Through a long-term evaluation of this trait, some rice germplasm involved in salt tolerance have been identified. More than 70 QTLs controlling the salt-related traits of Na+/K+ contents and survival days in rice have been identified. Two salt-tolerance genes SKC1 and DST have been cloned. A series of transgenic lines with salt-tolerance and polymerization lines of two salt-resistant genes (SKC1 and BADH) in rice have been developed in our lab. Finally, we discussed the prospects of rice salt-tolerant mechanism research and their applications in practice, which might provide an important reference for further studies of salt-tolerance in rice.
Salt-stress damage is one of the crucial factors contributing to unfavorable rice-production while soil salinization is the main limiting factor for directly influence its production (Li et al., 2005). Rice, planted in this kind of poor soil, often cut the production or even can not complete its life cycle when serious. Among 960 million hectares of saline-alkali soil worldwide, about 100 million hectares have existed in China. Rice fields suffering salt damage account for one fifth of all cultivated area in the continent. This salt damage can be reduced via managing soil and water, as well as modified chemical. However, it’s hard to come true for the high cost and its ineffectiveness. Therefore, it is one of the effective ways to ensure the food safety production of the salinization rice area and improve the ecological environment to cultivate salt-tolerant rice variety and do the salt tolerance research deeply in rice. By following those, it was done by the most botanists in China for a long time to protect the security of food and the environment (Zhang, 2005). In this paper, we summarized and prospected the germplasm resource, mechanism of salt-tolerance and corresponding molecular mechanism in rice, which might provide an important reference for further studies of salt- tolerance in rice.
1 The mechanism of salt-tolerance in rice
Salt tolerance refers to the ability of surviving and yielding while rice varieties cultivated in saline soil. When exposure to high-salt environment, plants cannot absorb water effectively, leading to a growth-delay, which is called the osmotic effect of salt stress. On the other hand, NaCl enters the respiratory chain so that the leaf cells accumulate large amount of Na+ which further impedes the growth of rice, which is called the ion effect of salt stress (Yang et al., 2008). The response of rice under salt-stress is as followings: maintaining the integrity of membrane system, ionic compartmentation, osmotic adjustment and accumulation of macromolecular protein etc.
1.1 Maintaining the integrity of membrane system
Under salt stress, the high salt solution first impairs the cell membrane by influencing the membrane’s permeability, composition and the role playing during transporting. These traits are obviously different when rice is cultivated in favorable conditions such as the increased permeability and the intensified peroxidation of membrane lipids, and these variations all obstruct normal physiological and biochemical functions of the plasma membrane, further affect the metabolism of the whole cell (Bing et al., 2008). Reactive oxygen including O2- and H2O2, can be produced in various ways such as the enzymatic reaction of plants, the electron transfer process of chloroplasts and mitochondria and the auto-oxidation of some low-molecular-weight organic compounds. However, they would be wiped out quickly by protective enzymes, saying SOD, POD, CAT and APX, leading to a physiological balance. POD, CAT and SOD constitute system of protective enzymes in plant. They worked in coordination to eliminate reactive oxygen produced by lipid peroxidation to protect the membrane structure, finally (Sun et al., 2008; Shu and Chen, 2000).
1.2 Ionic compartmentation
Salt-tolerate plants can resist or reduce the damage posed by salt-stress via regulating the ions absorption and compartmentation. Under this situation, the excessive ions accumulated in plants make many functional proteins and enzymes inactive leading up to a failure in cellular metabolism. However, plants do have a natural ability to protect themselves from injury produced by vast amount of inorganic ions Na+. Because most inorganic ions accumulated in cells are transported and stored in the vacuoles, thus plants can maintain normal growth in certain concentrations of salt solution, which is called the ionic compartmentation. Studies also show that this kind of compartmentation phenomenon exists in both strong and poor salt-tolerate plants, or even common plants, which illuminates that it might be a generally obtained ability in all plants, and it depends on the proton pumps located in membrane of vacuoles, they transport H+ to the outside of vacuoles from inside while Na+ was transported conversely. In this way, Na+ was accumulated in vacuoles. The power of transfer comes from gradient of proton concentration and electrochemical potential (Bing et al., 2008; Ren et al., 2005).
1.3 Plant osmotic regulation
Under the salt-stress, plants are easy to lose water and then are damaged badly. Due to the osmotic pressure of the external environment of high salt is low, it was forming a strong osmotic pressure difference between the plant internal environment and the external environment, which makes plants are difficult to absorb water, thus resulting in a phenomenon of water deficit. To avoid cell dehydration and maintain a stable water balance inside and outside cells, plant mainly increases the concentration of intracellular solute (small molecular soluble organics or inorganic ions) and decreases intracellular osmotic potential to regulate osmotic potential difference between cells. The osmotic adjustment of plant includes organic methods and inorganic methods. The first one involves proline, betaine, soluble carbohydrates, sugar alcohols and polyamines, etc. while the inorganic methods mainly involve inorganic ions such as Na+, K+, Ca2+ , Cl- and so on (Bing et al., 2008).
1.3.1 Ionic regulation
In order to maintain a stable micro-environment within cytoplasm, plant regulates the relative concentration of inorganic ions (mainly K+ and Na+) to adjust cell turgor, cell volume, intracellular pH value, ionic strength and many other crucial physiological parameters. Normally, in its entire physiological process, most plant cells accumulate K+ while transport Na+ outside to the cells leading to a high K+/Na+ rate across membrane, which is helpful for K+ to perform its unique functions. But, how do plant cells to maintain high K+/Na+ within cells? Studies have confirmed that two K+ absorption systems have existed on cell membrane—low-affinity K+ absorption system and high-affinity K+ absorption system. The low-affinity K+ absorption system is a K+ channel and K+ is transported through following a smooth concentration gradient from high to low. The high-affinity K+ absorption system is equipped with a symport-device and K+ can be transported against the concentration gradient depending on the energy produced by proton electrochemical gradient in the system. As a result, plant, under salt-stress situation, will transform low-affinity K+ absorption system to high-affinity system thereby enhancing the plant’s absorption of K+ and less accumulation of Na+. In this way, plant will survive longer under salt-stress condition with an improvement of salt-tolerance (Qi et al., 2007; Bing et al., 2008; Cheng et al., 2008).
1.3.2 Regulation of small organic molecular
Studies found that in high salt conditions, most plants began to accumulate some small molecular weight metabolites in cytoplasm such as proline, lycine, sugar alcohols and others to maintain high osmotic pressure within cells, making it possible for plants to absorb water under high-salt conditions with an increased stress resistance. As organic osmolytes, they must have some characteristics as follows: small molecular weight; soluble in water; without net charges at physiological pH; maintaining by membrane; little affection towards the structure of enzyme; quickly produced and accumulated enough contents to initiate osmotic regulation. In order to eliminate the imbalance caused by salt-stress, usually, plant accumulates two different types of substances to lower intracellular osmotic potential level and improve its salt tolerance. One kind of substance is small molecular organic compounds such as betaine, mannitol, sorbitol, proline and polyamines while the other is proteins including rice exogenous gene BADH, CMO, etc. (Shen et al., 2001; Guo et al., 1997).
2 Excavation of rice salt-tolerance germplasm
2.1 Identification methods of salt damage
Under the salt stress, rice plants usually present a variety of symptom response, such as hairy leaves, dehydration, no tiller, death, decrease of production, etc. This adaptability existed among different species, which presents various sensitivity to salt stress, is the foundation of salt-tolerance in rice germplasm resources.
There are three methods about identification of salt tolerance. (1) Salt resistance capacity of rice, which means that survival skills under the tough salinity- alkalinity stress, it is evaluated by biomass, vigor, etc. It can determine the potential and factual value in use under appointed tough environment, and it can also provide countermeasure of eco-cybernetics and basis of make rational use of rice resources. (2) Rice agricultural salt resistance capacity, which means rice agriculture productivity under salt stress, or rice yield under salt stress. It reflects the sensitivity of different varieties to salt stress. It can also determine the performance and adaptability of different varieties under appointed tough environment. (3) The response of isolated rice cell under salt stress, which is that the cell suspension cultures and callus of rice are cultivate under salt stress, and then compare the relative biomass or livability to determine salt-tolerance (Guo et al., 2003; Qi et al., 2005).
2.2 Rice salt-tolerant germplasm
The elite salt-tolerant germplasm is the carrier of genetic analysis of the salt tolerance and the breed of variety with resistance to salt stress. In the early 1930s, the breeders had launched to cultivate the salt tolerance rice variety. For example, the high salt tolerance native variety Pokkali which was cultivated in Sri Lanka; the salt tolerance commercial variety Kala Rata l-24 and hura Rata 4-10 were cultivated in India, the salt tolerance variety BRI, BR 203-26-2 and Sail were from Bengal. Since 1970, the International Rice Research Institute (IRRI) identified 10 salt tolerant varieties including Pokkali, Getu and Nona Bokra from 9 000 rice varieties and pedigree, which provided the technical reserve for the breed of the salt tolerance variety.
In China, the salt tolerance variety 80-85 was selected by the Academy of Agricultural Sciences in Jiangsu Province working with IRRI. The Chinese Academy of Agricultural Sciences selected 103 salt tolerance varieties including 27 indica, 76 japonica (the salt tolerance of some varieties were higher than Pokkali) from the 2808 introduced varieties, among which the 81-210, Nonglin72 and American rice achieved large area in coastal area of Jiangsu Province. Wu et al (1989, Jiangsu Agricultural Sciences, (l): 4-5) found some salt-tolerant germplasm such as Jiucaiqing, Laohuangdao, Huangjingnuo and Hongmangxiang- jingnuo from the native japonica in Taihu Lake Basin. China National Rice Research Institute has also screened out some salt tolerance varieties such as Mang Rice3, Hair Rice, Big Mang Rice and Sorghum rice (Table 1).
|
|
3 The genetic and molecular mechanism of the salt tolerance of rice
The salt tolerance of rice is the genetics of quantitative characters, which is controlled by multiple genes (Lin et al., 2004). The geneticists had mapped more than 70 QTLs which were related to the salt tolerance with the RIL and DH populations (Table 2), and two important salt tolerance genes of rice (SKC1 and DST) have already been cloned.
|
|
3.1 Mapping of the salt tolerance QTL of rice
The indicators of the salt tolerance mainly included the survival days in salt stress, salt damage level and Na+/K+ traits. So far, about 70 salt tolerance QTLs had been located (Zang et al., 2008). Lin et al (1998) detected one QTL (RG13) which was significant correlation with the salt tolerance on the chromosome 5 using the RIL population (Aishante 2/CB), and the locus could explain 11.6% of the phenotypic variances. Gu et al (2000) mapped 4 salt tolerance QTLs which affected the salt damage level and Na+ content in seedling with the BC1 population which was derived from the salt tolerance variety (Pokkali) and the salt sensitive variety (Peta), and found that 12 QTLs which affected the salt tolerance in mature stage distributed in 1 or 2 linkage interval of chromosome 7. Gong et al (Chinese Science Bulletin, 43(17): 1847-1850) mapped 7 QTLs of the survival days of seedling using the DH population which was derived from Zaiyeqing 8 and Jingxi 17 in 1998; he also investigated the differential expression of five important agricultural traits in the salt stress and the non-salt stress condition in 2000, and detected 17 QTLs (Gong et al., Science in China Series C, 30(6): 561-569). Koyama et al (2001) found 10 QTLs related to the Na+/K+ absorb of the stem and the leaf by employing the RILs population of IR4630/IR15324. Yao (2002) constructed the F2 population with Jiucaiqing and IR36 to detect the salt tolerance QTL in seedling stage by interval mapping, through which he reported 3 QTLs affecting salt damage level were located on chromosome 1, 5 and 9, respectively, 2 QTLs affecting Na+/K+ absorb of the root were located on chromosome 2 and 6, the QTLs affecting fresh weight of stem and leaf and the QTLs affecting dry weight of stem and leaf was mapped in the same interval, and located on chromosome 8 and 9, respectively, 2QTLs affecting the root length of the seedling were located on chromosome 4 and 5. Lin et al (2004) detected 11QTLs correlated with the survival days of seedling, Na+ /K+ absorb of the stem, the leaf and the root, which will pave the way for the further fine-mapping and cloning research.
3.2 Cloning of the salt tolerance gene of rice
SKC1: Ren et al (2005) isolated the SKC1, a major QTL for shoot K+ content, by map-based cloning, and found that it encoded a member of HKT-type transporters. They compared the SKC1 nucleotide sequences between a salt-tolerant indica Nona Bokra and a susceptible elite japonica Koshihikari allele, and found six nucleotide substitutions in the coding region which led to four amino-acid changes. Electro- physiological analysis showed that the protein encoded by SKC1 was specific transport proteins, but not transported directly for K+, while the variation of the content of K+ was caused by the competition of Na+. The analysis indicated that the protein located on the cell membrane. The report showed that the above ground of rice (leaf, stem) would accumulate a large number of Na+ in salt stress condition, while SKC1 could make the excess Na+ flow back to the root, which would reduce the toxicity of Na+ and enhance the salt tolerance of rice. There would be a wide application prospect of SKC1 in the molecular breeding of salt tolerance (Ren et al., 2005).
DST: A drought and salt tolerance (dst) mutant derived from a japonica cultivar Zhonghua 11 with ethylmethanesulfonate (EMS) treatment was identified, and the DST was cloned by the map-based cloning. DST encoded a previously unknown zinc finger transcription factor that negatively regulated stomatal closure by direct modulation of genes related to H2O2 homeostasis, which identified a novel pathway for the signal transduction of DST-mediated H2O2-induced stomatal closure. The assessment of the two amino acid substitutions (N69, A162) in the dst mutant showed that the N69 of DST was required for transcriptional activation. As a negative regulator, DST could directly down-regulated the expression of genes related to H2O2 metabolism when it lost the function, making the ability of removing H2O2 decrease, the accumulation of H2O2 in guard cell increase, then increases the stomata closure and reduces the water evaporation, consequently enhanced the salt tolerance of rice. Moreover, they found that the down-regulation of DST did not affect rice grain yield, which facilitated molecular breeding efforts to improve drought and salt tolerance in staple crops (Huang et al., 2009).
4 Breeding of salt tolerance rice
We have obtained significant progress on the research of the salt tolerance mechanism, the identification of the salt tolerant germplasm and the study of relevant molecular mechanism of rice, which will lay the foundation for the breeding of salt tolerance rice.
4.1 Direct using or conventional breeding of the salt tolerance germplasm of rice
To improve the salt tolerance of rice is not only to improve the yield, but also to expand the area of the saline soil. Breeders (Zhang et al., 2004; Yin et al., 2002) obtained a series of rice varieties with the resistance to different concentrations of salt using the known salt tolerance germplasm or through the conventional breeding, and the salt tolerance rice varieties like Liaoyan No.2, Dongnong 363, Changbai No. 6 and 7, Zhaiyeqing 8 and Tesanai No.2 had applied in production. In the slight saline soil, IRRI developed 8 t/hm2 of the production by using the salt tolerance variety without any improvement measures to the soil (Zhang et al., 2004).
4.2 Breeding of the salt tolerance of transgenic rice
Li et al (2005, Chinese Science Bulletin, 8: 613-617)) transferred the HAL2 gene (RHL) to the japonica Hejiang 19 with the via Agrobacterium-mediated transformation, the salt tolerance of the positive plants improved in the seedling stage, the injury of the cell membrane of the positive plants reduced, the vitality of the leaf enhanced and the salt tolerance increased. Zhi et al (2005) transferred the leguminous plant gene P5CS (2-hydrogen pyrrole 5-carboxylic acid synthase) into rice via the gene gun bombardment, and found that the content of the praline and the salt tolerance of transgenic rice also increased. Katsuhara et al (2003) transferred the gene encoded a water channel protein on the plasma membrane of the barley into rice and made it over-express, then they found that the ratio of the stem to the root in transgenic rice increased, while the sensitivity to salt stress of transgenic rice decreased. Li and Guo (2006) transferred the salt tolerance gene OPBP1 into rice via the gene gun bombardment, and the result showed that the transgenic plants grew fast, the chlorophyll content and biomass were significant higher than that of the control. Guo (1997) and Su (1999) transferred the single gene mtlD (1-phosphate mannitol dehydrogenase gene) and gutD (6-phosphate sorbitol dehydrogenase gene) or the bivalent gene CMO/BADH (choline monooxygenase gene/betaine aldehyde dehydrogenase base) into rice, and both all obtained the transgenic plants which the salt tolerance had enhanced.
Guo et al (2006) transferred the 5 genes related to salt tolerance CMO, BADH, mtlD (1-phosphate mannitol dehydrogenase gene), gutD and SAMDC (S-adenosylmethionine decarboxylase gene) into conventional japonica Xiushui 11, Zhonghua 11, indica Teqing and the restorer lines Minghui63 via Agrobacterium-mediated transformation and the gene gun bombardment, and combined with conventional hybridization breeding to polymerize pentavalent gene, then Xiushui 11 with 9 different genotypes were bred, among which the best line was selected, finally they got 9 combinations lines (8 single, bivalent, trivalent genes, and one pentavalent gene, the phenotypes all of these lines were similar to Xiushui 11, and the number were TX4CMO-4-13-6, TX4MtlD-5-9-12, TX4BADH-12-3-7, TX4GutD-14-9-7, TX4SAMDC-11-3-8, TX4CMO / BADH-10-3-9, TX4MtlD / gutD-7-9-14, TX4mtlD / gutD +SAMDC-16-4-9 and FX5 pentavalent -3-7), and the comprehensive assessment and the study of rational use were done in the southern part of Zhejiang Province.
4.3 The molecular breeding of two types of salt tolerance genes polymerization
Along with the development of genomics, gene pyramiding will be the most effective way to improve and cultivate the high-yielding rice varieties (Guo et al., 2008; Liu et al., 2003; Basharat et al., 2006). It will contribute to further improve the salt tolerance and breed the high salt tolerance variety to polymerize genes in different salt tolerance pathway. At present, two types of salt tolerance genes were used for breeding. One type was the genes CMO, BADH, mtlD, gutD and SAMDC, which was resistant to the water stress and regulate small-molecules like lycine; the other was anti ion toxicity gene SKC1 (Ren et al., 2005).
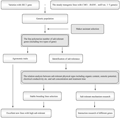
In 2005, we started to polymerize the above two types genes through combining the transgenic and the conventional breeding, which was based on the breeding of CMO / BADH transgenic rice and the marker-assisted selection of SKC1 resistant to salt tolerance, we wanted to obtain the salt tolerance rice with the genes in different salt tolerance pathway, to explore the genetic mechanism and interaction regulations of the two types genes and to improve the resistance to environmental stress and the commercial value of the genes (Figure 1). We had obtained a series of salt tolerance strains with the genes in different salt tolerance pathway by polymerizing two types of resistance genes molecular markers, which was funded by National 863, for example, Yueguang-SKC1 /BADH-12 and Xiushui 11-SKC1/BADH-23 could return to normal growth after treated in 1.0% NaCl. And the new stain Yuexiu T22-77 derived from transferring BADH/SKC1 into rice has already applied to the Ministry of Agriculture for intermediate experiment. The rice combinations polymerizing two types of resistance genes will process the wide resistance to environmental stress, the commercial value and application prospect.
|
|
5 Prospects
Plant salt-tolerance mechanisms have already been researched several decades and gotten many valuable achievements. This brings dawn to molecular breeding of plant salt tolerance, it also provides a possibility to combination of rice conventional breeding and molecular breeding (Gregorio et al., 2002; Garcia et al., 2004; Flowers, 2004). However, during the process of molecular breeding involved in salt tolerance by gene engineering, scientists have found some important questions which still were not clear. For instance, under salt stress, which factors were involved in accumulation of osmotic regulation substances? What are the first signal and transmitting process? What are the interaction and regulatory networks among several salt tolerance genes?
To solve the problem mentioned above, enhance the research of molecular mechanism involved in salt tolerance and application of production practice, the direction of research should be focused on various aspects, first of all, enhance the study of molecular mechanism involved in salt tolerance, this is the foundation of improve relevance and efficiency of molecular mechanism involved in salt tolerance. Secondly, combine multifunctional genes, major genes and minor genes. Rice salt tolerance is controlled by many genes, it is comprehensive performance of several physiological traits. As a result, during the research about salt tolerance of rice about physiological and gene engineering, more attention should be paid on synergistic action among several key factors. It is still unknown that the key factors which control rice salt tolerance. According to relevant background, isolation, cloning and genetic transformation of salt tolerance gene should carry on, in order to breed new varieties of salt tolerance rice by multi-gene plant expression and hybridization. Thirdly, QTL isolation and cloning of rice salt tolerance gene should carry on. After Ren (2005) and Huang (2009) get rice salt tolerance gene SKC1and DST separately according QTL Mapping, abundant QTL Mapping and cloning will be carried out. Fourthly, scientists should carry out innovative germplasm of rice salt tolerance actively. Transplanting the salt tolerance genes to rice improved breeds through conventional breeding, tissue culture, marker-assisted selection, genetically modification and gene polymerization and create salt tolerance rice species to satisfy urgent needs for high yield, good quality and salt tolerance rice. The last, exploit high saline-alkali tolerance rice from available resources, to provide abundant genes, more attention should be paid on indigenous rice and wild rice.
Some problems occurred during the research of salt tolerance breeding. For example, available cloned endogenous salt tolerance genes are seldom, and they are not closely connected with practice and application, as a result, they cannot produce great social and economic benefits; Biological security evaluation of exogenous gene transformation is still to be determined, and so on. But with the development of research and bio-technology, as well as recognition and support of the government to genetically modified rice, rice salt tolerance breeding will making great progress, in the near future, there will be huge number of salt tolerance rice varieties in production practice. It is of immense significance to relieve food supply crisis all around the world.
Authors’ Contribultions
SKH carried out the experiment; QQ and LBG conceived this program, directed the experimental design, data analysis, as well as manuscript writing and modification; HJT took part in the experiment and helped to analyze part of the data. All authors have read and approved the final manuscript.
Acknowledgement
This work was co-supported by National 863 Program Funded Projects (2006AA10Z1A9), State of Transgenic Special (2009ZX08001-022B) and the National Support Project (2006BAD13B01).
References
Basharat H.S., Ding X.H., Zeng L.X., Akshay T., Zhang Z.M., Zeng R.Z., and Zhang G.Q., 2006, Pyramiding four bacterial blight resistance genes into rice cultivars in south China, Molecular Plant Breeding, 4(4): 493-499
Bing L., Zhao B.C., Shen Y.Z., Huang Z.J., and Ge R.C., 2008, Progress of study on salt tolerance and salt tolerant related genes in plant, Journal of Hebei Normal University/ Natural Science Edition, 32(2): 243-248
Cheng Y.S., Yang H., Hou L.H., Chen Z.L., and Kang D.M., 2008, Study on integration, expression and salt resistance of three arabidopsis salt resistance genes in maize genome, Chinese Agricultural Science Bulletin, 24(2): 211-218
Flowers T.J., 2004, Improving crop salt tolerance, J. Exp. Bot., 55(396): 307-319
http://dx.doi.org/10.1093/jxb/erh003 PMid:14718494
Garcia A., Senadhira D., Flowers T.J., and Yeo A.R., 2004, The effects of selection for sodium transport and of selection for agronomic characteristics upon salt resistance in rice (Oryza sativa L.), Theoretical and Applied Genetics, 90(7-8): 1106-1111
Gong J.M., He P., Qian Q., Shen L.S., Zhu L.H., and Chen S.Y., 1998, QTL mapping of rice for salt tolerance, Kexue Tongbao (Chinese Science Bulletin), 43(17): 1847-1850
Gong J.M., Zhen X.W., Du B.X., Qian Q., Chen S.Y., Zhu L.H., and He P., 2000, Comparision of QTL controlling rice major agriculture characters between tolerance and no tolerance, Zhongguo Kexue C Ji (Science in China: Series C), 30 (6): 561-569
Gregorio G.B., Senadhira D., Mendoza R.D., Manigbas N.L., Roxas J.P., and Guerta C.Q., 2002, Progress in breeding for salinity tolerance and associated abiotic stresses in rice, Field Crops Research, 76(2-3): 91-101
http://dx.doi.org/10.1016/S0378-4290(02)00031-X
Gu X.Y., Mei M.T., Yan X.L., Zheng S.L., and Lu Y.G., 2000, Preliminary detection of quantitative trait loci for salt tolerance in rice, Chinese Journal of Rice Science, 14(2): 65-70
Guo L.B., Xue D.W., Wang H.Z., Chen S.Y., Lu D.Z., Zeng D.L., Gao Z.Y., Yan M.X., Huang D.N., and Qian Q., 2006, Improvement of rice salt-tolerance by using an integrated method of gene transformation and traditional breeding, Chinese Journal of Rice Science, 20(2): 141-146
Guo W.M., Fu Y.P., Sun Z.X., and Choung J.L., 2003, The correlation analysis between the morphological indices and salt tolerance in different rice germplasm under the salt stress, Journal of Plant Genetic Resources, 4(3): 245-251
Guo Y., Zhang L., Xiao G., Cao S.Y., Gu D. M., and Tian W. Z., 1997, Expression of betaine aldehyde dehydrogenase gene and salinity tolerance in rice transgenic plants, Science in China (Series C), 40(5): 496-501 http://dx.doi.org/10.1007/BF03183588 PMid:20229301
Huang X.Y., Chao D.Y., Gao J.P., Zhu M.Z., Shi M., and Lin H.X., 2009, A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control, Genes. Dev., 23(15): 1805-1817
http://dx.doi.org/10.1101/gad.1812409
Katsuhara M., Koshio K., Shibasaka M., Hayashi Y., Hayakawa T., and Kasamo K., 2003, Over-expression of a barley aquaporin increased the shoot/root ratio and raised salt sensitivity in transgenic rice plants, Plant and Cell Physiology, 44(12): 1378-1383
http://dx.doi.org/10.1093/pcp/pcg167 PMid:14701933
Koyama M.L., Levesley A., Koebner R.M., Flowers T.J., and Yeo A.R., 2001, Quantitative trait loci for component physiological traits determining salt tolerance in rice, Plant Physiol., 125(1): 406-422 http://dx.doi.org/10.1104/pp.125.1.406 PMid:11154348 PMCid:61021
Li B., Wang Z.C., Sun Z.G., Chen Y., and Yang F., 2005, Resources and sustainable resource exploitation of salinized land in China, Agricultural Research in the Arid Areas, 23(2): 154-158
Li N.Y., and Guo Z.J., 2006, Overexpression of two different transcription factors, OPBP1 and OsiWRKY, enhances resistance against pathogen attack and salt stress in rice, Chinese Journal of Rice Science, 20(1): 13-18
Lin H.X., Zhu M.Z., Yano M., Gao J.P., Liang Z.W., Su W.A., Hu X.H., Ren Z.H., and Chao D.Y., 2004, QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance, Theoretical and Applied Genetics, 108(2): 253-260
http://dx.doi.org/10.1007/s00122-003-1421-y PMid:14513218
Lin H.X., Yanagihara S., Zhuang J.Y., Senboku T., Zhen K.L., and Yashima S., 1998, Identification of QTL for salt tolerance in rice via molecular markers, Chinese Journal of Rice Science, 12(2): 72-78
Li R.T., Zhang Z.M., and Zhang Q.F., 2002, Salt tolerance of transgene plantlets and RHL transformation of non glutinous rice, Kexue Tongbao (Chinese Science Bulletin), 8: 613-617
Liu S.P., Li X., Wang Z.Y., Li X.H., and He Y.Q., 2003, Gene pyramiding to increase the blast resistance in rice, Molecular Plant Breeding, 1(1): 22-26
Qi D.L., Han L.Z., and Zhang S.Y., 2005, Methods of characterization and evaluation of salt or alkaline tolerance in rice, Journal of Plant Genetic Resources, 6(2): 226-231
Qi D.L., Guo G.Z., Lee M.C., Cao G.L., Zhang J.G., Zhou Q.Y., Zhang S.Y., Suh S.C., and Han L.Z., 2007, Progress of physiology and genetic research on saline-alkaline tolerance in rice, Journal of Plant Genetic Resources, 8(4): 486-493
Ren Z.H., Gao J.P., Li L.G., Cai X.L., Huang W., Chao D.Y., Zhu M.Z., Wang Z.Y., Luan S., and Lin H.X., 2005, A rice quantitative trait locus for salt tolerance encodes a sodium transporter, Nat. Genet., 37(10): 1141-1146
http://dx.doi.org/10.1038/ng1643 PMid:16155566
Shen Y.G., Du B.X., Zhang J.S., and Chen S.Y., 2001, Cloning and characterization of CMO gene from atriplex hortensis, Chinese Journal of Biotechnology, 17(1): 1-6
Shu W.G., and Chen S.Y., 2000, Gene expression and signal transduction in plants under osmotic stress, Progress in Biotechnology, 20(3): 3-6
Su J., Chen P.L., and Ray W., 1999, Transgene expression of mannitol- 1-phosphate dehydrogenase enhanced the salt-stress tolerance of the transgenic rice seedlings, Scientia Agricultura Sinica, 32(6): 101-103
Sun J.C., Wang X.S., and Yang S.L., 2008, Progress of research on salt-resistance in plants, Agricultural Research in the Arid Areas, 26(1): 226-230
Wang B., Lan T., and Wu W.R., 2007, Mapping of QTLs for Na+ content in rice seedlings under salt stress, Chinese Journal of Rice Science, 21(6): 585-590
Wu R.S., Wang Z.X., Jiang H., Wang G.L., Gu P., Chen B.Q., 1989, Screening and identification of rice salt tolerance genetic resources in Taihu lake valley, Jiangsu Agricultural Sciences, (l):4-5
Yang X.H., Peng X.J., Yang G.H., Feng L.L., Wang K., and Li Y.S., 2008, Preliminary function analysis of rice OsRab7 in salt tolerance and construction of its genetic transformation vector, Journal of Wu han Botanical Research, 26(1): 1-6
Yao M.Z., 2002, Inheritence analysis of salt tolerance of a rice landrace jiucaiqing (Japonica) from taihu valley, Thesis for M.S., Nanjing Agrieultural University, Supervisor: Zhai H.Q., pp:43-46
Yin Y.L., Liang J.S., Liu Q.Q., Wang Z.G., and Liu C.X., 2002, Cloning of HAL1 gene from saccharomyces cerevisiae and construction of it’s plant expression vector, Journal of Yandzhou University (Agricultural and Life Science Edtion), 23(4): 27-29
Zang J.P., Sun. Y., Wang Y., Yang J., Li F., Hou Y.L., Zhu L.H., Jessica R., Mohammadhosein F., Xu J.L., and Li Z.K., 2008, Dissection of genetic overlap of salt tolerance QTLs at the seedling and tillering stages using backcross introgression lines in rice, Science in China Series C: Life Sciences, 51(7): 583-591
http://dx.doi.org/10.1007/s11427-008-0081-1 PMid:18622741
Zhang G.H., Zeng D.L., Guo L.B., Liu H.J., Hu J., Gao Z.Y., Hua Z.H., and Qian Q., 2007, Nutrition-functional rice created by polymerizing ADP-glucose pyrophosphorylase (AGP) and giant embryo (ge) genes, Chinese Journal of Rice Science, 21(6): 567-572
Zhang J.W., Tan X.L., and Xu J., 2004, Effect on seeding of various rice strains of different concentions of NaCl solution, Journal of Plant Genetic Resources, 5(1): 100-104
Zhang X.Q., 2005, Studies on salt tolerance of transgenic rice, Zhongguo Daomi (China Rice), 6: 10-12
Zhi L.F., Chen M.Q., Yu T., Cao J.W., Zhu Y.G., and Li Y.S., 2005, The study of the transformation by p5cs in rice cells, Journal of Hubei Normal University (Natural Science), 25(4): 39-43
. PDF(233KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Shikai Hu
. Hongjian Tao
. Qian Qian
. Longbiao Guo
Related articles
. Rice
. Salt-tolerance
. Germplasm
. QTL
. Genetics and breeding
Tools
. Email to a friend
. Post a comment
.png)
.png)