 Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2020, Vol. 11, No. 4 doi: 10.5376/rgg.2020.11.0004
Received: 15 May, 2020 Accepted: 18 May, 2020 Published: 28 Aug., 2020
Chen Y., Liang Y.L., Du S.L., and Zhang Y., 2020, Identification and gene mapping of a white panicle mutant wp7 in rice, 11(4): 1-8 (doi: 10.5376/rgg.2020.11.0004)
Lots of leaf color mutants have been reported in rice, while the panicle color mutants were less. The principle of development mechanism and photosynthesis of the panicle chloroplasts need further study (Sun et al., 2015). White panicle mutant of rice plays an important role in the study of rice functional genome and light morphogenesis, and also has an important application prospect in seed preparation and variety improvement (Zhang et al., 2013). In the reported white panicle mutant materials at present, the leaf usually show albino or white stripes at seedling stage, and the degree of albino and panicle phenotype are also different.
Among the fully albino mutants in the panicle, the plants died when the degree of albino of wp1 was severe, while white stripes were distributed on the leaves from seedling stage to flowering stage when the albino was light. At heading stage, panicle had albino phenotype, WP1 located on chromosome 7 (Sanchez and Khush, 1994). It encoded a Val-tRNA synthetase OsVALRS2, which regulated ribosome biosynthesis in chloroplasts (Wang et al., 2016). wp2 seedling leaf was white striped, reverted to green after 4-leaf, and the glume, flower axis and pedicel were all albino after heading, and the mutation gene was located on chromosome 6 (Sanchez and Khush, 2000). slwp leaves had white stripes with panicle albino from 2-leaf stage to the heading stage, and the gene was located in the region of 0.91 Mb chromosome 6. Sequencing analysis showed that the gene encoded RNRS1 is a ribonucleotide reductase small subunit protein (Zhou et al., 2018), which is likely to be a pair of alleles with STWP (Chen et al., 2015) and WLP6 (Li et al., 2018). Fully albino panicle mutants have great influence on yield and other agronomic traits, which is not conducive to breeding and reproduction. Among the mutants with incomplete albino panicle and white-striped leaves at young leaf stage, the wp3 mutants with white palea, lemma, cob, and part with green filamentous stripes. WP3 localized on chromosome 1, encoding a mitochondrial protein, and regulated the chloroplast development in rice panicles by maintaining functional mitochondria (https://doi.org/10.3389/fpls.2018.00762). After the 3-leaf stage, wsp1 appeared white green stripe on the vein, and the leaf color of seedling and panicle was obviously abnormal. The plastid in the white part of leaf was abnormal, without any lamellar structure. The mutant gene was located on the chromosome 4. Wsp1 encoded a family protein of organelle RNA editing factor (MORF) with high homology with Arabidopsis MORF2 (Zhang et al., 2017). wslwp seedlings had white stripes, but after 2-leaf stage, it gradually reverted to green from the tip of the leaf along the vein to fully recover at the mature stage. After heading, outer glume albino, but the cob and branches were normal. The gene was located in 87 kb of chromosome 7 (Jin et al., 2011). Such mutations will not cause rice seedling death and yield reduction. In the mutant materials with panicle albino and normal leaf, the wp4 leaves were primrose yellow, the chloroplasts of inner and outer glumes could not complete normal development, the glumes were milky white, while the cob was green, and the gene was located in the range of about 79 kb on chromosome 8 (Wang et al., 2015). The leaf of wp6 mutant was normal, the glumes remained albino after heading, the branches and cob were green, and there was no report on its location (Cong et al., 2018). These mutants have no obvious changes in yield and other agronomic traits, could be used as morphological markers in rice-cross, and improved seeds breeding. Besides, there are also temperature-sensitive mutants: Leaf of the low temperature-sensitive mutant wlp1 albino at seedling stage and panicle albino at heading stage with low temperature 23℃. WLP1 gene is located on chromosome 1 and is a coding gene of 50 ribosomal L13 proteins on chloroplasts (Song et al., 2014). Two allelic wlp2s and wlp2w were found in the high temperature-sensitive mutant WLP2, all of which were albino lethal at high temperature. WLP2 encodes a PEP-associated protein. WLP2 and its paralog OsFLN2 can physically interact with thioredoxin OsTRXz to form a TRX-FLN regulatory module, which not only regulates transcription of the PEP-encoded genes but also maintains the redox balance in chloroplasts under heat stress, participate in the development of rice chloroplast biogenesis under heat stress, and protect the chloroplast development from heat stress (Lv et al., 2017). All of the above mutations were controlled by a pair of recessive nuclear genes. While st-fon was cytoplasmic inheritance, and white streaks appeared on its culm, leaves and panicles, the number of floral organs increased and florets cracked and could not be closed (Chen et al., 2013).
In this study, we found a new white panicle mutant in rice, similar to wp6 (Cong et al., 2018). There was no significant difference with the wild type at the seedling stage, but the difference was genetic analysis. It showed that the white panicle character of rice was controlled by two recessive genes, which was quite different from the reported white panicle mutant. The morphological identification, determination of photosynthetic indexes and pigments in panicles, chloroplast ultrastructure observation, genetic analysis and gene mapping of wp7 were carried out. It is of great significance to clone and verify the related genes, clarify the mechanism of regulation of panicle chloroplast development, and increase the understanding of photosynthetic mechanism of non-leaf tissues.
1 Results and Analysis
1.1 Exophenotype of white panicle wp7
White panicle material wp7 was not significantly different from normal materials at seedling stage. At heading stage, white panicle phenotype appeared in wp7 (Figure 1A). wp7 displayed albino glume with green branches and cob. In the same panicle, there were few glumes fully albino palea and lemma and concentrated at the top of the panicle (Figure 1B). According to the degree of albino of wp7 glume, it could be divided into three categories: albino palea and lemma (Minimum, 2.9%), green stripe palea and albino lemma (Maximum, 63.0%), green stripe palea and lemma (Middle, 34.1%). The above results indicated that gene mutation had a great influence on the color of glume.
.png) .png) Figure 1 Comparison of plant, panicle, and glumes appearance between 18A8 (Normal) and wp7 at heading stage Note: A: Plant phenotype, Bar=20 cm; B: Panicle characters, Bar=2 cm; C: Glume phenotype (a: Albino palea and lemma; b: Green stripe palea and albino lemma ; c: Green stripe palea and lemma), Bar=2 mm |
1.2 Glume chloroplast structure of white panicle wp7
In order to find out the cytological reasons of glume albino, the wp7 white part, green part and chloroplast of normal green panicle were observed and compared by TEM (transmission electron microscope). The chloroplasts of normal green panicle were distributed in the parenchyma cells of the glume (Figure 2A). The lamellar structure of chloroplasts was clear, regular and large (Figure 2B; Figure 2C; Figure 2D). Observed the green part (Figure 2E; Figure 2F; Figure 2G; Figure 2H) and white part (Figure 2I; Figure 2J; Figure 2K; Figure 2L) of the wp7 panicle, we found that the chloroplast of green part was normal (Figure 2F), the starch granules were much, the grana was less and small (Figure 2G) and the lamellar structure was sparse and fuzzy (Figure 2H). In the white part, we only found a few, small and abnormally developed plastids, and the electron density in the matrix was low (Figure 2I; Figure 2J). There were osmiophilic granules, no thylakoid lamellae (Figure 2K), and appeared large vacuoles in some plastids (Figure 2L). The results showed that gene mutation caused serious damage to the structure of glume chloroplast.
.png) .png) Figure 2 Effects of wp7 mutation on chloroplast number and structure in white panicle wp7 Note: A~D: Wide type green panicle; E~H: wp7 white part; I~L: wp7 green part; C: Chloroplast; G: Grana; M: Mitochondria; O: Osmiophilic granules; P: Plastid; S: Starch granule; A,E,I: Scale = 5 µm; B,F,J: Scale = 2 µm; C,G: Scale = 1 µm; K,L: Scale = 200 nm; D,H: Scale = 100 nm |
1.3 Determination of chlorophyll content and photosynthesis in panicle of mutant wp7
In order to find out the change of chlorophyll content in the panicle of wp7, we determined the chlorophyll content of white and normal green panicles after heading. The results showed that the content of chlorophyll a and total chlorophyll in white panicle wp7 were lower than those in normal green panicle, and the difference was significant (p<0.05) (Figure 3). The contents of chlorophyll b and carotenoid (c) were low, and the difference was not significant. The above results indicated that the chlorophyll synthesis function of the mutant glume was seriously lost, which is mainly reflected in the significant decrease of chlorophyll a content.
.png) .png) Figure 3 Determination of chlorophyll content of wp7 glume Note: Ca: Chlorophyll a; Cb: Chlorophyll b; Ca+b: Total chlorophyll; Cc: Carotenoids; *: Significant difference (p<0.05) |
In order to know the effect of mutation on panicle photosynthesis, photosynthetic indexes of wp7 and 18A8 panicle were determined in the field. The results showed that the net photosynthetic rate (Pn) of wp7 was significantly lower than that of normal plants. The stomatal conductance (Gs) and intercellular CO2 concentration (Ci) of WP7 were higher than that of normal plants. And there was no significant difference in transpiration rate (Tr) (Table 1). It indicated that the mutation seriously affected the photosynthetic performance of panicle.
.png) Table 1 Measurement of Photosynthetic Indicators Note: The data are shown as means±SD from 3 individual replicates; *: Significant difference (p<0.05) |
1.4 Genetic analysis
In order to clear the genetic rule of white panicle wp7, F1 and F2 generations were produced by crossing wp7 with normal green panicle 18A8. The F1 generation and backcross BC1F1 generation were produced by crossing sanming dominant male sterility (sms) (green panicle) with wp7. All F1 plants had normal green panicles. F2 and BC1F1 generations were separated, and the separation ratio of normal green panicle and white panicle plants was 15:1 in the cross-combination of F2 and 18A8. The separation ratio of sms and wp7 backcross generation (BC1F1) was 3:1 (Table 2). The results showed that the albino phenotype of wp7 mutant may be controlled by two pairs of recessive genes with overlapping effects.
.png)  Table 2 Separation ratio of F2 and BC1F1 |
1.5 Gene mapping of mutant wp7
From the backcross population of rice (sms/wp7//wp7), 12 white panicle and 12 normal individuals were selected to form a gene pool, and the mutation site of wp7 was located. The results showed that among 351 InDel molecular markers in rice genome, 02005 primer (Table 3) had polymorphism between white panicle plant and normal green panicle plant. Genotypes of some green panicle and white panicle plants were analyzed with primer 02005. The results showed that the marker was linked to glume color (Figure 4).
.png) 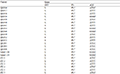 Table 3 Primer sequence of polymorphism markers |
.png) .png) Figure 4 Validation of linkage between the glume color and marker 02005 Note: ▽: Recombinants |
In order to locate the target gene between two molecular markers, 20 pairs of new primers were designed on both sides of 02005 to screen new polymorphic markers and genotypes. Finally, we found that the five new InDel markers C02D081, C02D092, C02D084, C02D089, and C02D095 (Table 3) showed polymorphic among their parents, which were all linked to glume color by individuals verification (Figure 5).
.png) .png) Figure 5 Genotyping of InDel marker C02D095 Note: ▽: Recombinants |
Furthermore, the genotypes of 187 white panicle individuals were analyzed by these five polymorphic markers, and 37, 37, 4, 3 and 29 recombinants were detected respectively. According to the genetic distance between the markers and the white panicle traits and the inclusion relationship among the recombinants, the wp7 related genes were mapped between the markers C02D089 and C02D095. The genetic distances between the markers and the genes were 1.6 cM and 15.5 cM, respectively (Figure 6). Because genetic analysis showed that there may be two genes related to the white panicle, but we only located one site, so we named the site WP7-1.
.png) .png) Figure 6 Linkage map of WP7-1 |
2 Discussion
wp7 only showed albino in the glume, the branches and cobs were normal green, glumes were not completely albino, some panicle had green stripes on the pelea and lemma. From the top to the base of the panicle, the degree of albino of the glumes in wp7 decreased gradually, and the proportion of albino lemma and green stripe palea was the largest. Only 15 white panicle mutants have been reported, and wp7 has enriched rice white panicle resources. There was no obvious abnormality in leaf color of wp7, but only albino in the panicle, which was similar to the reported white panicle materials wp4 (Wang et al., 2015) and wp6 (Cong et al., 2018). Meanwhile, the albino glume of wp7 showed green stripe, were not completely albino, which was similar to the wp4 (Wang et al., 2015), St-fon (Chen et al., 2013) and wp3 (https://doi.org/10.3389/fpls.2018.00762). They were stripe mutants. The similarity of chloroplast ultrastructure between wp7 and this kind of stripe mutants is that there are most normal chloroplasts in green part of glume and a few abnormal chloroplasts which lack lamellar structure and unclear inner membrane. Moreover, there was no chloroplast in the white part of wp7. The plastid became smaller with abnormal shape. And there was no normal lamellar structure in the later stage. Many vesicular structures appeared in some plastids.
WP7-1, mapped in this study, was located on chromosome 2, between InDel Markers C02D089 (physical position 19444169) and C02D095 (physical position 21337678). WP7-1 did not recombine with the reported mutant gene wp4 between SSR marker RM2-97 and RM13553 on chromosome 2. Therefore, there may be a new white panicle gene at wp7-1 locus. In order to further understand the mechanism of panicle photosynthesis, it is necessary to make fine mapping and cloning.
The genetic analysis of white panicle wp7 indicated that the white panicle trait might be controlled by two pairs of recessive nuclear genes, but only one marker linked to the white panicle trait was found. One of the reasons is that the white panicle trait of wp7 was only controlled by one recessive gene. However, the white panicle mutant may die due to its low vitality, which cannot normally pass through the stages of germination and seedling, resulting in the decrease of white panicle individuals and the low proportion of white panicles in the mapping population. The other reason is that the white panicle of wp7 is controlled by two recessive genes, but the linkage markers of wp7 cannot be screened for there is no polymorphism near another locus or the polymorphism of primary screening primers is poor. In order to determine the number and precise location of wp7 white panicle genes, one is to select high-quality near isogenic lines to construct a single locus mapping population, so as to avoid the mutual interference between genotype and phenotype in the identification process. The other is to screen more polymorphic markers or construct larger mapping population with large genetic differences parents to determine another locus and narrow the mapping interval.
3 Materials and Methods
3.1 Experimental materials
White panicle parent wp7: Sirio. Normal panicle parent: 18A8 and sms (provided by Huang xianbo, Sanming Academy of Agricultural Sciences). F1: sms/wp7, 18A8/wp7. F2: 18A8/wp7 selfing. Backcross F1: sms/wp7//wp7.
3.2 Observation on the phenotype of mutants
After heading, the plant morphology and glume color of wp7 and 18A8 were compared by ordinary camera and zoom-stereo microscope.
3.3 Observation on the chloroplast
At heading stage, 1×1×1 mm3 glume was selected and fixed with 2.5% glutaraldehyde, vacuumed for 3 times, washed with PBS (0.1 mol/L, pH 7.2) for 3 times, fixed at 4℃ with 1% osmic acid, and then washed with PBS (0.1 mol/L, pH 7.2) for 3 times again. After that, it was dehydrated at 30%, 50%, 70%, 80%, 90%, 95%, 100% (twice) ethanol and acetone gradients, and then embedded in epoxy resin at room temperature and cured overnight at 60℃. Ultrathin sections were cut on a copper mesh and stained with uranyl acetate and lead citrate, and then observed by transmission electron microscopy (Type of FEI AIG2SPIRIT TWIN).
3.4 Determination of chlorophyll content and photosynthetic index
At heading stage, 0.1 g new glume of panicle was weighed, and then cut into pieces and added into test tube. Chlorophyll content and carotene content were measured according to the reference (Wellburn, 1994). Repeated 5 times and took the average value.
At heading stage, 5 green panicles and 5 white panicles plants were measured by LI-6400 portable photosy- nthetic analyzer from 9:00 am to 11:00 am on a sunny day. The cluster leaf chamber was selected under the following conditions: CO2 concentration 340±10/cm3/m3, light quantum flux 1 000±50 μmol m-2s-1 (Wei et al., 2003), and then the actual measured part was cut off to measure the panicle surface area (Teare and Peterson, 1971), and the recalculation function was selected to obtain the back calculation data.
3.5 Genetic analysis
At heading stage, the glume color of F1 and F2 of mutant wp7×18A8, F1 generation of sms×wp7 and backcross F1 were observed and the segregation ratio was calculated.
3.6 Gene mapping
The mixed DNA of 15 green panicles and 15 white panicles plants in BC1F1 population was extracted by CTAB to construct two gene pools, which were diluted 30 times as template for screening polymorphism among gene pools. The primers were 351 Indel marker primers located on 12 chromosomes of rice. The mapping of genes and linkage markers was constructed according to the method provided by Michelmore et al. (1991) and Zhang et al. (2008).
Authors' contributions
CY and LYL designed and carried out this experiment. CY and LYL participated in the data analysis and drafted the manuscript. LSL participated in the experiment. ZY conceived of the study, and guide its design, data analysis, draft, and revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 31370349).
Chen D.X., Li T., Qu G.L., Huang W.J., He Z.Q., and Li S.G, 2013, Genetic analysis of streaked and abnormal floret mutant st-fon, Rice Sci., 20(4): 267-275
https://doi.org/10.1016/S1672-6308(13)60132-8
Chen Y.P., Zhai Z., Yang W.J., Sun J., Shu X.L., and Wu D.X., 2015, Genetic analysis and fine mapping of St-wp gene in mutant rice with stripe white leaf and white panicle, Henongxue Bao (Journal of Nuclear Agricultural Sciences), 29(7): 1246-1252
Cong X.H., Ruan X.M., Luo Z.X., Bai Y.S., Yang L.S., Luo Y.X., and Shi F.Z., 2018, Discovery and study of white panicle mutant wp6 in Rice, Anhui Nongye Kexue (Journal of Anhui Agricultural Sciences), 46(36): 29-31
Jin Y., Liu H.Q., Wang D.K., and Tao Y.Z., 2011, Genetic analysis and gene mapping of a white striped leaf and white panicle mutant, Zhongguo Shuidao Kexue (Chinese Journal Rice Science) 25(5): 161-166
Li L.F., Xiong Y.Y., Ouyang L.J., Peng X.S., Chen X.R., He X.P., Fu J.R., Bian J.M., Hu L.F., Xu J., He H.H., Sun X.T., and Zhu C.L., 2018 Identification and gene mapping of white stripe leaf and white panicle mutant wlp6 in rice, Zhongguo Shuidao Kexue (Chinese Journal Rice Science), 32(6): 538-548
Lv Y.S., Shao G.N., Qiu J.H., Jiao G.A., Sheng Z.H., Xie L.H., Wu Y.W., Tang S.Q., Wei X.J., and Hu P.S., 2017, White leaf and panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice, J. Exp. Bot., 68(18): 5147-5160
https://doi.org/10.1093/jxb/erx332
PMid:29045742 PMCid:PMC5853965
Michelmore R.W., Paran I., and Kesseli R.V., 1991, Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations, Proc. Natl. Acad. Sci. USA, 88(21): 9828-9832
https://doi.org/10.1073/pnas.88.21.9828
PMid:1682921 PMCid:PMC52814
Sanchez A.C., and Khush G.S., 1994, Chromosomal location of some marker genes in rice using the primary trisomics, J. Heredity, 85(4): 297-300
https://doi.org/10.1093/oxfordjournals.jhered.a111461
Sanchez A.C., and Khush G.S., 2000, Chromosomal localization of five mutant genes in rice, Oryza sativa, using primary trisomics, Plant Breed., 119(1): 84-86
https://doi.org/10.1046/j.1439-0523.2000.00424.x
Song J., Wei X.J., Shao G.N., Sheng Z.H., Chen D.B., Liu C.L., Jiao G.A., Xie L.H., Tang S.Q., and Hu P.S., 2014, The rice nuclear geneWLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions, Plant Mol. Biol., 84(3): 301-304
https://doi.org/10.1007/s11103-013-0134-0
PMid:24132771
Sun Y.L., Li W.C., and Ji S.D., 2015 Research overview on the rice seeding leaf chlorosis genes, Hubei Noneye Kexue (Hubei Agricultural Science), 54(11): 2564-2568
Teare I.D., and Peterson C.J., 1971, Surface area of chlorophyll-containing tissue on the inflorescence of Triticum aestivum L.1, Crop Sci., 11(5): 627-628
https://doi.org/10.2135/cropsci1971.0011183X001100050006x
Wang Y.L., Wang C.M., Zheng M., Lyu J., Xu Y., Li X.H., Niu M., Long W.H., Wang D., Wang H.Y., Terzaghi W., Wang Y.H., and Wan J.M., 2016, WHITE PANICLE1, a Val-trna synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development, Plant physiol., 170(4): 2110-2123
https://doi.org/10.1104/pp.15.01949
PMid:26839129 PMCid:PMC4825129
Wang X.W., Jiang Y.D., Liao H.X., Yang B., Zou S.Y., Zhu X.Y., He G.H., and Sang X.C.,2015, Identification and gene fine mapping of white panicle mutant wp4 in Oryza sativa, Zuowu Xuebao (Acta Agronomica Sinica), 41(6): 838-844
https://doi.org/10.3724/SP.J.1006.2015.00838
Wei A.L., Wang Z.M., Zhai Z.X., and Gong Y.S., 2003, Effect of soil drought on c4 photosynthetic enzyme activities of flag leaf and ear in wheat, Zhongguo Nongye Kexue (Scientia Agricultura Sinca), 36(5): 413-417
Wellburn A.R., 1994, The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution, J. Plant Physiol., 144(3): 307-313
https://doi.org/10.1016/S0176-1617(11)81192-2
Zhang Y., Li Y.F., Zhang J., Shen F.C., Huang Y.X., and Wu Z.W., 2008, Characterization and mapping of a new male sterility mutant of anther advanced dehiscence t in rice, J. Genet. Genomics, 35(3): 177-182
https://doi.org/10.1016/S1673-8527(08)60024-7
Zhang Z.G., Cui X.A., Wang Y.W., Wu J.X., Gu X.F., and Lu T.G., 2017, The RNA editing factor WSP1 is essentia for chloroplast development in rice, Mol. Plant, 10(1): 86-98
https://doi.org/10.1016/j.molp.2016.08.009
PMid:27622591
Zhang H.Z., Cheng Z.J., and Wan J.M., 2013, Progresses on the studying of rice leaf albino, Shengwu Jishu Tongbao (Biotechnology Bulletin), 11: 1-7
Zhou K.N., Xia J.F., Ma T.C., Wang Y.L., and Li Z.F., 2018, Mapping and mutation analysis of stripe leaf and white panicle gene SLWP in rice, Zhongguo Shuidao Kexue (Chinese Journal Rice Science), 32(4): 325-334
. PDF(468KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yun Chen
. Liang Yanling
. Du Shuanglin
. zhang yi
Related articles
. Rice ( Oryza sativa )
. White panicle
. Chloroplast
. Gene mapping
Tools
. Email to a friend
. Post a comment


