Research Report
Introgressing Blast Resistant Gene Pi-9(t) into Elite Rice Restorer Luhui17 by Marker-Assisted Selection 
2. Luzhou Branch of National Rice Improvement center, Luzhou, 646100, P.R. China
 Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2011, Vol. 2, No. 4 doi: 10.5376/rgg.2011.02.0004
Received: 31 Oct., 2011 Accepted: 07 Nov., 2011 Published: 16 Jan., 2012
Wen et al., 2011, Introgressing Blast Resistant Gene Pi-9(t) into Elite Rice Restorer Luhui17 by Marker-Assisted Selection, Rice Genomics and Genetics, Vol.2, No.4 31-36 (doi: 10.5376/rgg.2011.02.0004)
In this research, we introgressed the broad-spectrum blast resistant gene Pi-9(t) from the donor parent P2 into hybrid restorer Luhui17 by using backcross approach and molecular marker-assisted selection (MAS) technique. We detected the target gene through the specific molecular marker pB8 that close linked to the blast resistant gene Pi-9(t). As a result, sixty eight lines carrying broad-spectrum blast resistant gene Pi-9(t) were obtained, and then four lines, WR1023, WR1043, WR1056 and WR1062, were identified to be homozygous backcross lines with resistant gene Pi-9(t) by molecular phenotyping identification. Furthermore, the lines, WR1023 and WR1056 lines, were proved to have genetically stable agronomic characteristics, that demonstrated good plant architecture, high blast resistance and stable restoring ability, Therefore we suggested that these lines should be used as resistant gernplasms in in rice hybrid breeding program.
Rice blast is one of the three diseases in rice crop, usually leading to completely yield loss in the region of the serious incidence. More than 80 countries around the world have reported that blast occurred, leading to the loss of yield each year nearly 10 million tons of rice product. In China blast damage led to the yield loss of each year more than one hundred million kilograms (Baker et al., 1997, Dong et al., 2000). As the variations of pathogenicity of physiological races occurs more frequently, so some resistant varieties after cultivated 3 or 5 years often gradually lost the resistance to rice blast. Developing blast resistant varieties would be still the most cost-effective method to improve rice blast disease resistance in rice. To date, more than 80 blast resistant genes have been mapped, of which Pib, Pita, Pid2, Pi9, Piz-t, Pi36 and Pi37 has been isolated and cloned (Wu et al., 2007; Qu et al., 2006; Lin et al., 2007). Pi-9(t), one of the identified blast resistant genes, exhibited excellent resistance to rice blast by identified with rice blast races from 43 different countries (Liu et al., 2002), which was considered as the widest spectrum resistant resource favored by the breeders in the cloned rice blast resistant genes. The resistance of rice varieties to rice blast was mostly controlled by a pair or several pairs of main effective dominant genes. The achievements have been made in applications of the blast resistant genes in rice blast resistant breeding program, such as the Pid1, Pib, Pita pyramided to G46B (Chen et al., 2004), the Pi2 introduced into Zhenshan 97B (Chen, 2004) and the Pi1, Pi2, Pi33 23B introgressed to Jin 23B (Chen et al., 2008). A batch of new varieties were developed exhibiting that the resistant abilities to rice blast were significantly improved by field identification.
Traditional rice breeding for disease resistance mainly depends on the selections of field resistant performance and plant trait phenotypes, being subject to influences of subjective factors as well as environmental factors. Traditional breeding being longer selection cycles often happens in the loss of resistance genes in the selection process. Therefore, the breeders have to have extensive field experiences due to less selective efficiency. Marker-assisted selection is a kind of breeding selection for target genes through analysis on molecular markers closely linked to the genotype of the target gene. Because of its selective effects being independent from gene effects and environmental factors, the selection results were considered as being higher reliability and genotype selection could be performed in early generation, thus speeding up the breeding program (Chen et al., 2005).
In this study, Luhui No.17, a restorer of super rice â…¡ You No.7, as maternal parent and P2 donor parent containing Pi-9(t) gene were employed to make cross and backcross, screening new restorer carrying Pi-9(t) gene in the separating offspring by combining the use of marker-assisted selection and identification of field resistance induced by artificial inoculation, thereby enhancing the resistance of restorer and new hybrid rice combinations to rice blast in order to realize the comprehensive breeding objectives of high yield, good quality and disease resistance.
1 Results and Analysis
1.1 To obtain Homozygous lines containing the Pi-9(t) gene with excellent agronomic traits
The F1 plants were obtained by crossing of susceptible Luhui17 (R17) as maternal parent and P2 as donor parent, then using Luhui17 (R17) as recurrent parent generated BC1F1 and BC2F1 by backcrossing with two times. Molecular markers pB8 closely linked of Pi-9(t) gene was employed to perform marker-assisted selection in the beginning of BC1F1 generation to choose individuals containing with Pi-9(t) gene and similar agronomic traits with Luhui17 (R17) for further backcrossing. Target gene of each generation needs to be identified by in-door PCR. The improved individuals containing blast resistant gene Pi-9(t) screened from the separating generation of the BC2F2 population were continuous self-crossing and finally the target lines with genetic stability were applied to be identified in field blast resistant validation and comparative trials of agronomic traits (Figure 1).
|
Figure 1 Breeding procedure for developing rice lines with Pi-9(t) gene |
1.2 4 Homozygous lines containing Pi-9(t) gene identified by molecular markers and agronomic traits
Molecular markers pB8 tightly linked to Pi-9(t) gene used to detect the polymorphisms of donor parent P2 recurrent parental LuHui17 (R17), the results showed in figure 2 that molecular marker pB8 could generate about 500 bp of characteristic band in donor parent P2, while no band in recurrent parent Luhui17 (R17), indicating that the tightly linked molecular markers pB8 should exhibit obvious polymorphism between donor and recipient parents, so this marker can be used in marker-assisted selection. The separating offspring were detected by PCR amplification using molecular markers pB8 to present one banding pattern in the F1 lines with the same as the donor parent P2. Whereas polymorphic markers pB8 used to detect 116 individuals of BC2F2 population to present 68 individuals with the same banding pattern as P2, and 48 individuals without band like Luhui17 (R17) (Figure 2). The results showed that pB8 can effectively detect the different genotypes in the separating generation eventually to breed favorable lines with Pi-9(t) gene and excellent agronomic traits. By use of this marker continue to monitor the target gene in the separating generations as well as to identify resistance to rice blast and favorable traits, eventually four lines containing homozygous Pi-9(t) gene and superior agronomic traits came out in the generation of BC2F3 (Figure 3).
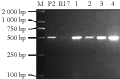 Figure 2 Polymorphism among Luhui17, P2 and their segregating population BC2F2 detected by pB8 |
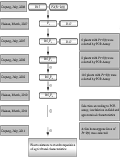 Figure 3 Four homozygous lines containing Pi-9(t) gene identified by PCR |
1.3 Resistant identification of improved lines and their combinations to rice blast
Field resistant trials to rice blast were carried out in the disease nursery in Pujiang County of Sichuan Province in 2010 and 2011. Nursery disease identification in 2010 showed that the leaf blast scores of 4 lines carrying Pi-9(t) gene with the background of restorer Luhui17, WR1023, WR1043, WR1056 and WR1062, decreased compared with the control Luhui17, of which the leaf blast scores of WR1043, WR1056 and WR1062 decreased three grades from 7 to 4, except for WR1023 maintaining the same leaf blast score as the control Luhui17. Incidence rate of neck blast decreased from 100% to 18%~32% while spike blast scores decreased down to 5 from 7. It was obvious that the resistance to rice blast was significantly improved. Likewise Nursery disease identification in 2010 showed that the leaf blast scores of 4 lines carrying Pi-9(t) gene with the background of restorer Luhui17, WR1023, WR1043, WR1056 and WR1062 17, decreased compared with the control Luhui17, of which the leaf blast scores of WR1043, WR1056 and WR1062 decreased three grades from 8 to 3 or 4 (Table 1). Incidence rate of neck blast decreased from 85% to 16%~28% while spike blast scores decreased down to 5~7 from 9. The improved resistant effects were still significant. The above two years results of resistant identification showed that the two-year trend of rice blast incidence of restoring lines carrying Pi-9(t) gene were basically identical, of which the rice blast resistant scores of three new candidate lines had far more markedly risen than that of the recipient parent, while close to the resistance of donor parent.
|
Table 1 Rice blast resistance identified for parents and four candidate lines in the 2010 and 2011 |
1.4 Agronomic evaluation of candidate lines
The main agronomic traits of candidate lines were compared with their the parents (Table 2), exhibiting super-parent phenomenon in plant height with significant differences from parental receptor. There was no any significant difference in the trait of the effective panicles per plant, average panicle length, full filled seeds, the number of seed setting per panicle, seed setting rate and kilo-grain weight compared to parental receptor.
|
Table 2 Performance of agronomic traits of candidate restoring lines and their controls |
1.5 Agronomic evaluation of combinations between candidate restorer and â…¡-32A
Agronomic traits of combinations between candidate line and CMS â…¡-32A (Table 3) were compared with the control, exhibiting the significant differences in plant height, the numbers of full filled grains and seed-setting rate while no significant difference in the traits of the effective panicles per plant, average panicle length, the number of grain per panicle and kilo-grain weight. Comprehensive analysis of agronomic traits showed that the combinations with WR1023 and WR1056, two improved candidate restorers presented better performance in the traits of effective panicles per plant, full filled grains per panicle and seed setting rate. Because of the candidates with own excellent agronomic traits, the tests for the combining ability would be more extensive carried out to screen the optimal combinations for continuing variety trials or provincial trials.
|
Table 3 The agronomic performances of rice hybrids by combining candidate restoring lines with â…¡-32A and controls |
2 Discussion
In this study, we tried to develop novel resistant materials to rice blast by genetically improving the rice blast resistance of Luhui17, an elite backbone restorer of hybrid rice. Blast resistance creates new materials. The hybrid combinations by restore line of Luhui17 have been applied in the rice production, possessing lots of superior characters, such as high seed production etc. However, the application of these hybrid combinations in rice production has been limited because of losing rice blast. In this study, we obtained new lines containing broad-spectrum blast resistant gene Pi-9(t) and restore gene by molecular marker-assisted selection and conventional breeding methods. As the blast resistant gene Pi-9(t) is the dominant resistant gene, it will facilitate to enhance resistant ability of hybrid combinations to rice blast that would continuing contribute to food production in China.
In recent years, the technology of molecular marker-assisted selection has made great progresses in resistant breeding program, for example, the photosensitive genic male sterile 3418s (Luo et al., 2003), rice restorer lines of R8006 and R1176 (Cao et al., 2003) and Kang 4183 (Luo et al., 2005) were successfully developed with a high resistance to bacterial blight by using the bacterial blight resistant gene xa21 introduced into rice CMS restorer line. The rice blast resistant gene Pid1, Pib, Pita were pyramided to G46B (Chen et al., 2004), the rice blast resistant gene Pi2 introduced into Zhenshan 97B (Chen et al., 2004), and the rice blast resistant genes Pi1, Pi2, Pi33 were pyramided into Jin 23B (Chen et al., 2008), of which the resistance of these enhanced lines and their combined hybrids have greatly improved. In this study, we obtained resistant individuals containing Pi-9(t) gene and four restore lines with Pi-9(t) gene and restore gene in BC2F3 though the approaches of marker-assisted selection and field identification of agronomic traits. Identification of resistance in the field trials showed that the resistant levels of the new lines have markedly risen, of which was higher than the recipient parent as well as close to the donor parent. Our results were quite similar to previous studies. Two lines named WR1023 and WR1056, being stability and good plant architecture, high resistance to rice blast and high combining ability, could be directly used in rice production or in resistant breeding program. It is no doubt that using marker-assisted selection approaches could greatly shorten the breeding period, elevate breeding efficiency as well reduce the cost of breeding, which has been exhibited great application prospect.
3 Materials and Methods
3.1 Rice materials used in this research
Rice materials used in this research including: (1) P2 as donor parent, partial japonica intermediating materials derive from the indica- japonica cross containing Pi-9(t) gene, came from the International Rice Research Institute. (2) Luhui17 (R17) as receptor parent, excellent restore line of the super rice â…¡ you No.7 with high combining ability, without resistance to rice blast.
3.2 Identification of rice blast
Blast resistance was identified using the approach of natural disease-induced nursery in the station of rice blast disease nursery in Pujiang County of Sichuan Province. Each rice material were planted two rows, 10 individuals per row with row space 16 cm row spacing and 16 cm plant inter- spacing were transplanted individually, surrounded by planting disease induced material. Conventional field management was applied without any pest control. Field disease surveying, recording and resistant level scoring were followed the evaluation criteria titled The standard technical specifications Y/T of rice blast resistance of agricultural industry of the People's Republic of China (http://www.ccvi.net/trial/200311106.htm). In this study, disease grade of leaf blast and panicle blast as well as the incidence rate of neck blast were scored.
3.3 DNA extraction and PCR analysis
DNA extraction was followed with the Ni's method (Ni et al., 2005) with slightly modification. Taking fresh young rice leaves about diameters of 4 cm into the 1.5 mL eppendorf tube, adding liquid nitrogen to grind into powder and rapidly adding 600 μL extract buffer (100 mmol/L Tris-HCl pH 8.0, 20 mmol/L EDTA, 500 mmol/L NaCl, 1.5% SDS), incubating at 65℃ water bath for 30~40 min, then adding 600 μL mixture of chloroform and isopentanol with 24 to 1 and mixing at room temperature for standing 30 min. After centrifuging at 10 000 r/min for 5 min, the supernatant was transferred to another centrifuge tube, then adding an equal volume of ethanol for blending, discarding the supernatant after centrifugation prior to standing for 1 h, while precipitation was washed with 70% ethanol 2 times, then adding 600 μL sterile water for fully dissolving after naturally dried, placed in refrigerator at 4℃ ready for use.
Two primers for pB8 were designed based on the sequence of molecular markers pB8 tightly linked to the reported Pi-9(t), including P1: 5'-CCGGACTAAGTACTGGCTTCGATA-3' and P2: 5'-CCCAATCTCCAATGACCCATAAC-3' (Liu et al., 2002). In 25 μL PCR reaction system were contained : 10×PCR Buffer 2.5 μL; 25 mmol/L MgCl2 3 μL, 2 mmol/L dNTPs 2 μL, Taq polymerase (5 U/μL) 0.5 μL, primers of P1 (25 pmol/L) 1 μL and P2 (25 pmol/L) 1 μL, total genomic DNA 2 μL and adding ddH2O 13 μL up to total volume. PCR amplification procedure was followed as pre-denaturation at 94℃ for 5 min, then 30 cycles of denaturation at 94℃ for 1 min, annealing at 55℃ for 1 min and extension at 72℃ for 1.5 min, finally extension at 72℃ for 10 min. PCR products were separated by 1.5% agarose gel electrophoresis in the 90 constant voltage for 2 hours, the bands were observed and scored in gel imaging system.
3.4 Field trials and agronomic trait phenotyping
Field trails were carried out in the years of 2010 and 2011 at the Deyang station of Rice Sorghum Institute, Sichuan Academy of Agricultural Sciences, planted four stable lines with Pi-9(t) gene, Luhui17, the donor parent P2 and four combinations by crossing four tested lines and CMS line â…¡-32A. Random block design with three replicates for each materials was arranged and 3 rows with total 30 individuals each replicates were planted with the spacing of 16.5 cm×26.4 cm. Five individuals was sampled in each plot for phenotyping. Plant height, panicles per plant, single panicle length, grains per panicle, and full filled seed per panicle, seed setting rate, and kilo-grain weight and so on were investigated.
Authors' contributions
Shaoshan Wen is the principal investigator who was responsible for project and conceived experimental design of this study as well as wrote the manuscript. Shaoshan Wen and Bijun Gao both are the executor of the experiment, data analysis and paper revision. All authors have read and agree the final context.
Acknowledgements
This study was jointly funded by Youth Foundation of Sichuan Academy of Agricultural Sciences (2009QNJJ-30), Special fiscal funded breeding project of Sichuan Province, (2007YZGC11-31), Sichuan Science and Technology Support Program Deyang City, (2011NZ027) and Key Technology Plan project of Luzhou city in Sichuan.
References
Baker B., Zambryski P., Staskawicz B., and Dinesh-Kumar SP., 1997, Singaling in plant-microbe interactions, Science, 276(5313): 726-733
http://dx.doi.org/10.1126/science.276.5313.726 PMid:9115193
Cao L.Y., Zhuang J.Y., Zhan X.D., Zheng K.L., Cheng S.H., 2003, Hybrid rice resistant to bacterial blight developed by marker assisted selection, Zhongguo Shuidao Kexue (Chinese Journal of Rice Science), 17(2): 184-186
Chen H.Q., Chen Z.X., Ni S., Zuo S.M., Pan X.B., and Zhu X.D., 2008, Pyramiding Three Genes with Resistance to Blast by Marker-Assisted Selection to Improve Rice Blast Resistance of Jin 23B, application, Zhongguo Shuidao Kexue (Chinese Journal of Rice Science), 22(1): 23-27
Chen X.W., Li S.G., Ma Y.Q., Li H.Y., Zhou K.D., and Zhu L.H., 2004, Marker-assisted selection and pyramiding for three blast resistance genes, Pi-d(t)1, Pi-b, Pi-ta2, in rice, Shengwu Gongcheng Xuebao (Chinese Journal of Biotechnology), 20(5): 708-714
Chen Z.W., Guan H.Z., Wu W.R., Zhou Y.C., and Han Q.D., 2005, The screening of molecular markers closely linked to rice blast resist an t gene Pi-1 and their application, Fujian Nonglin Daxue Xuebao (Journal of Fujian Agriculture and Forestry University (Natural Science Edition)), 34(1): 74-77
Cheng Z.W., Zheng Y., Wu W.R., and Zhao C.J., 2004, Screening and Application of an SSR Marker Closely Linked to Pi-2(t), a Gene Resistant to Rice Blast, Fenzi Zhiwu Yuzhong (Molecular Plant Bleeding), 2(3): 321-325
Dong J.X., Dong H.T., and Li D.B., 2000, Recent advances on rice blast disease resistance, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 8(1): 99-102
Lin F., Chen S., Que Z.Q., Wang L., Liu X.Q., and Pan Q.H., 2007, The blast resistance gene Pi37 encodes a nucleotide binding site-leucine-rich repeat protein and a member of a resistance gene cluster on rice chromosome 1, Genetics, 177(3): 1871-1880
http://dx.doi.org/10.1534/genetics.107.080648 PMid:17947408 PMCid:2147969
Liu G., Lu G., Zeng L., and Wang G.L., 2002, Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6, Mol. Genet. Genomics, 267(4): 472-480
http://dx.doi.org/10.1007/s00438-002-0677-2 PMid:12111554
Luo Y.C., Wang S.H., Li C.Q., Wang D.Z., Wu S., and Du S.Y., 2003, Breeding of the photo-period-sensitive genetic male sterile line ‘3418S’ resistant to bacterial bright in Rice by molecular marker assisted selection rice, Zuowu Xuebao (Acta Agronomica Sinica),29(3): 402-407
Luo Y.C., Wang S.H., Li C.Q., Wang D.Z., Wu S., and Du S.Y., 2005, Improvement of bacterial blight resistance by molecular marker-assisted selection in a wide compatibility restorer line of hybrid rice, Zhongguo Shuidao Kexue (Chinese Journal of Rice Science), 19(1): 36-40
Ni D.H., Yi C.X., Li L., Wang X.F., Wang W.X., and Yang J.B., 2005, Pyramiding Xa21 and Pi9(t) in rice by marker-assisted selection, Fenzi Zhiwu Yuzhong (Molecular Plant Breeding), 3(3): 329-334
Qu S.H., Liu G.F., Zhou B., Bellizzi M., Zeng L., Dai l., Han B., and Wang G.L., 2006, The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multi-gene family in rice, Genetics, 172(3): 1901-1914
http://dx.doi.org/10.1534/genetics.105.044891 PMid:16387888 PMCid:1456263
Wu J., Liu X.L., Dai L.Y., and Wang G.L., 2007, Advances on the identification and characterization of broad-spectrum blast resistance genes in rice, Shengming Kexue (Chinese Bulletin of Life Sciences), 19(2): 233-238
. PDF(634KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Shaoshan Wen
. Bijun Gao
Related articles
. Rice ( Oryza sativa L.)
. Blast resistance
. Pi-9(t)
. Molecular marker-assisted selection
. Rice restorer
Tools
. Email to a friend
. Post a comment


