Molecular Phylogeny of Indian Indigenous Aromatic Rice Based on Sequence Diversity of the Chloroplast-encoded matK Gene 
 Author
Author  Correspondence author
Correspondence author
Rice Genomics and Genetics, 2015, Vol. 6, No. 8 doi: 10.5376/rgg.2015.06.0008
Received: 30 Jul., 2015 Accepted: 11 Sep., 2015 Published: 22 Sep., 2015
Patil R.G., Jadhao K.R., Samal K.C., and Rout G.R., 2015, Molecular Phylogeny of Indian Indigenous Aromatic Rice Based on Sequence Diversity of the Chloroplast-encoded matK Gene, Rice Genomics and Genetics, Vol.6, No.8 1-8 (doi: 10.5376/rgg.2015.06.0008)
Phylogenetic relationships were inferred using nucleotide sequences of the chloroplast gene matK for 10 aromatic rice cultivars and its wild relatives along with the progenitor’s grasses families. Multiple sequence alignment (MSA) of 10 genotypes revealed higher number of substitution between the 1110 bps to 1270 bps which might be the crucial during evolution of aromatic rice. Similarly MSA of all related sequences with aromatic rice cultivars showed substitution and deletion in aromatic rice cultivars. The deletion of ‘T’ at position 1761 and 1769 was crucial nucleotide changes in aromatic rice as compared to its progenitors. The aligned sequences were used for molecular phylogenetic analysis by neighbor-joining methods with 1000 replication of bootstrap test. Phylogenetic tree of aromatic rice cultivars showed cultivar ‘Banikunja’ was outgroup whereas cultivars ‘Basumati dhan’ and ‘Gatia’ was more closely related. Similarly phylogenetic tree of aromatic rice with its wild relatives and grasses progenitors showed that aromatic rice was more closely related to Oryza sativa japonica group. The genus Oryza was divided into two main clades and evolved from completely different group of grasses families. Oryza brachyantha has high affinity for grasses and should be treated as a progenitor for wild Oryza species.
Introduction
Aromatic rice constitute a small but special group of rice, which are considered best for aroma and have occupied a prime position in society for aroma and cooking qualities (Ahuja et al., 1995). A large number of aromatic rice has already been disappeared and many are at the verge of extinction. Therefore, there is a utmost necessary to conserve the germplasm. The chloroplast maturase K gene (matK) is one of the most variable coding genes of angiosperms and has been suggested to be a “barcode” for land plants. A 'barcode' gene that can be used to distinguish between the majorities of plant species on earth has been identified. However, matK exhibits low amplification and sequencing rates due to low universality of currently available primers and mononucleotide repeats. In plant systematic, matK has recently emerged as an invaluable gene because of its high phylogenetic signal compared with other genes used so far (Muller et al., 2006).The 1500 bp matK gene is nested in the group II intron between the 50 and 30 exons of trnK in the large single copy region of the chloroplast genome of most of the green plants (Sugita et al., 1985; Steane, 2005; Daniell et al., 2006; Turmel et al., 2006). Phylogenetic analysis of a data set composed of matK, rbcL, and trnT-F sequences from basal angiosperms demonstrated that matK contributes more parsimony informative characters and significantly more phylogenetic structure on an average per parsimony-informative site than the highly conserved chloroplast gene rbcL (Muller et al., 2006). Sequence information from matK alone has generated phylogenies as robust as those constructed from data sets comprised of 2-11 other genes combined (Hilu et al., 2003). Further, the molecular data generated from matK has been used to resolve phylogenetic relationships at taxonomic levels (Johnson and Soltis, 1994; Hayashi and Kawano, 2000; Hilu et al., 2003; Cameron, 2005). The matK gene stands out among plastid genes used in plant systematic for its distinct mode of evolution. The rate of substitution in matK is three times higher at the nucleotide level and six times higher at the amino acid level than that of rbcL which denoting it as a rapidly evolving gene (Johnson and Soltis, 1994; Olmstead and Palmer, 1994; Soltis and Soltis, 2004). Various scientist have been studied the plant evolution, and able to solve various anomalies in the taxonomic levels by using the chloroplast genes such as matK and rbcL. The matK gene has two unique features that emphasize its importance in molecular biology and evolution. It is characterized by its fast evolutionary rate and putative function as a group II intron maturase. It is a chloroplast encoded gene nested between the 5’ and 3’ exons of trnK, tRNA-lysine in the large single copy region of the chloroplast genome (Sugita et al., 1985).
Molecular sequence data has revolutionized evolutionary studies and enhanced the revolution of phylogenetic trees immensely. Genes used in plant systematic display different trends of evolution. Slow evolving genes such as rbcL and atpB, have high sequence conserve among plant groups. This high sequence conservation allows a good resolution that has been confined to the family level, but cannot solve the intricacies below this level (Goldman et al., 2001). The matK gene is considered to be fast evolving due to the fact that it has a high rate of substitution and more variable sites compared to other genes (Olmstead and Palmar, 1994). Some of the researchers reported that the matK has been considered as a pseudogene because they contain stop codons within the ORF, bear indels that create frame-shift mutations and display an equal level of substitution for all three codons position (Kores et al., 2000). Additionally, frame-shift mutations found in the 3’ region of matK of the family Poaceae, which could also alter or destroy the reading frame; appear to be limited to the very 3’ region of this gene, not affecting the functionally of domain X (Hilu and Alice, 1999).
The genus Oryza, which includes rice and closely related wild relatives, has emerged as a powerful system to study the modes and mechanisms of genome evolution (Ammiraju et al., 2008). The genus Oryza comprises approximately 23 species that have been grouped into six diploid (AA, BB, CC, EE FF, GG) and four allotetraploid genome types (BBCC, CCDD, HHJJ and KKLL) (Lu et al., 2009). All Oryza polyploids are wild and contain important phenotypic traits that have the potential for use to improve the cultivated rice (Brar and Khush, 1997). In the present study implies the understanding of major evolutionary relationships of aromatic rice under the family, genus and species level by using the chloroplast derived matK gene.
Results and Discussion
Among the various approaches used in molecular systematic, DNA sequencing has become one of the most widely utilized, particularly above the genus level. Sequence variation in the coding and spacer regions of a number of chloroplast, mitochondrial, and nuclear genes has been utilized, with some genes more widely used, like the matK, Adh1, Adh2 and rbcL, and both coding and intergenic spacer region (ITS) of the ribosomal DNA (Clegg et al., 1994; Hsiao et al., 1994; Clark et al., 1995). The sequence data are cumulative, the potential sizes of informative data sets are immense, and the data are available in public computer databases. The matK gene is emerging gene as potential contributions to plant molecular systematic and evolution (Johnson and Soltis, 1994, 1995; Steele and Vilgalys, 1994; Liang and Hilu, 1996). This gene has been used effectively in addressing systematic questions in the number of families i.e. Polemoniaceae (Johnson and Soltis, 1995), Poaceae or Gramineae (Liang and Hilu, 1996), Orchidaceae tribe Vandeae (Jarrell and Clegg, 1995) and Myrtaceae.
In this investigation, sharp and bright bands of 1500 kbs of plastid matK gene fragment from template DNA of ten aromatic rice varieties were amplified by using matK specific primers (Figure 1). For the sequencing large scale amplification was done and amplified fragments were separated in 2.5% low agarose gel electrophoresis and subsequently used for elution though Gel Extraction Kit (GeneiTM). The ten eluted fragments of rice varieties were sequenced with an ABI 3730 XL genetic analyser with a BigDye terminator cycle sequencing kit (SciGenome, Kerala, India). For correctness of the sequences, DNAs isolated from independent varieties were analyzed for each genotype.
All nucleotide sequences from ten aromatic rice genotypes were subjected to multiple sequence alignment (MSA) in Muscle program. The conserved region was identified in all aromatic rice genotypes and found very little changes in the conserved region. With close observation of all ten genotypes sequences, it was found that the sequences of all genotype were muted during evolution and showed number of changes such as substitution, insertion and deletion in the cultivars (data not given). The number of substitution was higher between the 1110 bps to 1270 bps which might be the crucial during evolution. Similarly, Kron et al. (1999) was also found the insertion and deletion in matK of Epacrids and vaccinioids. Meanwhile MSA of ten aromatic rice genotypes along with wild relatives of rice and grasses revealed that the conserved nucleotide sequences as well as nucleotides changes during evolution of aromatic rice as compared to wild relatives and grasses under family Poaceae. The key mutational changes such as substitution ‘G’ to ‘C’ and ‘A’ was found in cultivars except ‘Banikunja’ and ‘Basaomati (Paikanapua)’ viz. ‘Basnasapuri’, ‘Basnaparijat, ‘Basumati dhan’,‘Chatiamaki - 1’, ‘Dhoiabankoi’, ‘Ganjam local - 2’, ‘Gatia’ and ‘Kalikati - 1’ at position 1352 bp respectively. Whereas, most variable nucleotide sequences was present in all aromatic rice genotypes at position 1389 to 1384 (Figure 2A) and complete deletion of nucleotide ‘T’ was observed at position 1761 and 1769 in all ten aromatic rice genotypes (Figure 2B). Relevant to these issues is also indicating that a gene is not a homogenous population of nucleotides since different parts of the gene assume different functional responsibilities and some sections might lack a function. Consequently, rates of nucleotide substitution can vary along the entire coding, or even the noncoding regions of a gene (Sastri et al., 1992; Clegg et al., 1994; Clark et al., 1995).
|
Figure 1 Amplification of 10 aromatic rice cultivars employing matk gene specific primer. Note: Ladder= Low range DNA ruler plus; Name of cultivars represents in the lower of the gel; Numbers on the right side of margin represents molecular weight marker DNA in base pairs (bps) and Number on left side represents size of amplified fragments in bps |
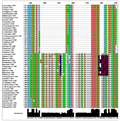 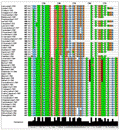 Figure 2 Jalview of muscle multiple sequence alignment tool (www. ebi.ac.uk/ Tools/msa/ muscle/) based on nucleotide sequences of 10 aromatic rice cultivars and its wild relatives along with grasses progenitors. Note: A: Colored boxes in the MSA indicate the substitution nucleotide changes in aromatic rice cultivars; B: Red colored boxes indicates deletion of nucleotides with respect to its progenitors. |
Phylogenetic tree for matK gene based on nucleotide sequences for both aromatic rice genotypes along with wild relatives and grasses under family Poaceae based on the alignment dataset was constructed by neighbour joining statistical method of bootstrap test with 1000 bootstrap replications in MEGA 6. Phylogenetic tree of ten aromatic rice genotypes showed one outgroup and two sub-clusters. Genotype ‘Banikunja’ was an outgroup, whereas genotypes ‘Basnasapuri’, ‘Ganjam local-2’, ‘Basumati dhan’, ‘Gatia’ and Basnaparijat’ were placed in cluster-IA and ‘Dhoiabankoi’, ‘Chatiamaki-1’, ‘Basaomati (Paikanapua)’ and ‘Kalikati-1’ was in cluster-IB (Figure 3). Divergence time scale from ‘Basnasapuri’ to ‘Banikunja’ was estimated which was found maximum for genotype ‘Banikunja’ i.e 0.015 and minimum for ‘Gatia’ and ‘Dhoiabanki’ i.e. 0.009. The maximum 47 and minimum 9 bootstrap values were observed. Evolutionary divergence between the sequences were estimated by using pair wise distance matrix for aromatic rice cultivars it was found that genotypes of ‘Banikunja’ and ‘Ganjam local-2’ was distantly related with maximum divergence value of 0.0296. whereas, Basumati dhan and Gatia was closely related with minimum divergence value of 0.0190 (Table 1).
 Figure 3 Phylogenetic tree based on nucleotide sequences of 10 aromatic rice cultivars constructed by neighbor joining method Note: Out group indicates in red mark. Black bold fond on upper side of the tree node indicates bootstrap values in per cent. Values in black fond below the tree root indicates the divergence time |
Phylogenetic analysis of aromatic rice along with related sequences was performed in order to find out the relationship between aromatic rice, wild relatives and grasses under family Poaceae in MEGA 6 program. Phylogenetic analysis of 41 other rice species and grasses along with ten aromatic rice genotypes were subjected to study the evolutionary pattern of aromatic rice. It observed that there are two main groups evolved from common ancestor. Phylogenetic tree was categorized into two main groups cluster-I and cluster-II. Cluster-I was further divided into two sub-clusters IA and IB and aromatic rice genotypes was placed in sub sub-cluster IA1 which indicates the aromatic rice was evolved after cultivated rice viz. Oryza sativa indica and japonica group. Similarly cluster-II was also divided into two sub clusters IIA and IIB. The 12 wild relatives of Oryza group was completely separated during evolution and placed differently in sub sub-cluster- IIB1 (Figure 4). These 12 wild relatives of Oryza group might be evolved from closely related grasses such as Maltebrunia letestui, Prosphytocloa prehensilis and Leersia oryzoides. Whereas domesticated rice Oryza sativaindica group, Oryza sativa japonica group and its wild relatives were evolved from another group of grasses such as Leersiatis seranti, Chikusichloa aquatica, Potamophila parviflora, Rhynchoryza subulata and Zizania aquatica. Phylogenetic tree revealed that aromatic rice was evolved from Oryza sativa japonica group and Oryza sativa indica group along with the parallel evolution of Oryza glaberrima and Oryza rufipogal. Similarly, Khush (2000) was also suggested that the common rice and the African rice, Oryza glaberrima was an example of parallel evolution in crop plants. Oryza nivara was the possible ancestor of Oryza sativa indica and japonica group. The maximum bootstrap value 99 was calculated for Cluster-II including Oryza ridleyi and Oryza longiglumis which indicates the correctness of topology and minimum bootstrap value 1 was estimated for Oryza rufipogal which indicated the interior branch.
|
Figure 4 Phylogenetic tree based on the alignment of nucleotide sequences ofaromatic rice cultivars and its wild relatives along with progenitors grasses constructed by neighbor joining method. Note: Common rice indicates in red and green mark. Black bold |
In the conclusion, this study indicates that the patterns and types of nucleotide substitutions, insertion and deletion in the matK gene are most comprehensive phylogenetic analysis using molecular sequence data. The phylogenetic study of the matK nucleotide sequences of ten aromatic rice genotypes including related wild species and other grasses family under Poaceae concludes that the genus Oryza was divided into two main clades and evolved from completely different group of grass families. Aromatic rice might be evolved from Oryza sativa (japonica and Indica group), Oryza glaberrima or Oryza rufipogal because of its low bootstrap value support. Oryza brachyantha has high affinity for grasses and should be treated as a progenitor for wild Oryza species. The study will also address the issues of optimal number of nucleotides essential for robust phylogenies and the consequences of utilizing different segments of the gene.
Materials and Methods
Genotypes
Ten promising upland as well as lowland cultivars of indigenous aromatic rice (Oryza sativa L.) viz. ‘Basnasapuri’, ‘Basnaparijat, ‘Basumati dhan’, ‘Banikunja’, ‘Basaomati (Paikanapura)’, ‘Chatianaki - 1’, ‘Dhoiabankoi’, ‘Ganjam local - 2’, ‘Kalikati - 1’, and ‘Gatia’ were collected from the Rice Research Station, Orissa university of Agriculture and Technology, Bhubaneswar, Odisha, India and used in the present study.
DNA extraction and PCR amplification
Total cellular DNAs were extracted from young leaftissues of ten aromatic rice genotypes using modified CTAB method (Edwards et al., 1991) and purified the DNA of each genotype was subjected for PCR amplification using overlapping oligos viz. matKF1: 5’TAATTAAGAGGATTCACC AG 3’ and matKR1: 5‘ATGCAACACCCTGTTCTG AC3’ (Merck Bioscience, India) by examining the previous study on rice (Ge et al., 1999).PCR amplification was carried out with the template DNA (25~50 ng), 2.5 µl 10X PCR assay buffer (Merck Bioscience, India),1.5 µl each of 10 mM dNTPs (M/S Merck Bioscience, India), 1 µl of 5 µM forward primer, 1 µl of 5 µM primer and 1 µl of 1U Taq DNA polymerase (Merck Bioscience). M/s Peqlab, 96 universal gradient thermal cycler was used for the PCR amplification consisted ofa total of 35 cycles of initial denaturation (94°C for 4 min) followed by denaturation (94°C for 1 min), annealing (57°C for 2 min), elongation (72°C for 2 min) and final elongation (72°C for 15 min). The 1 X Tris-acetate ethylene diamine tetra acetic acid (TAE) buffer was used to electrophoreses PCR amplified products in 2.5% (w/v) agarose gel (Merck Bioscience, India) along with 3 kb ladder (Himedia Laboratories Pvt. Ltd., Mumbai). The gel image was documented using gel documentation system (UVITECH, Cambridge, UK).
PCR product purification and sequencing
Large-scale amplification was performed to elute bright PCR fragment. The single bright amplicon of 1500 kb was eluted using Gel Extraction Kit (GeneiTM).The PCR amplified DNA fragments were subjected for two way sequencing in order to get the full sequence and minimize the error in sequencing and was sequenced by using both 96 capillary high throughput sequencer; ABI 3730 XL system to generate sequences with accurate base calling which is an extension and refinement of Sanger’s dideoxy method (Sanger et al., 1977) at SciGenom, Cochin, Kerala, India.
Sequence Data Analysis
The sequenced nucleotide datasets were manually edited in BioEdit program (version 5.0.9 http://www. mbio.ncsu.edu/BioEdit/bioedit.html) and used for further analysis. Alignment of all ten sequences were toggled using Multiple Sequence Alignment (MSA) of muscle programme (www. ebi.ac.uk/Tools/MSA/ muscle/) and alignments were visualized in Jalview. Meanwhile BlastN program was used for preparation of dataset based on the alignment results of nucleotide for all related sequences were downloaded from the GeneBank database and analyzed in MSA along with ten aromatic rice cultivars, MEGA (Molecular Evolutionary genetic analysis) version 6 was used for phylogenetic analysis and calculations of pair wise distances [http://www.megasoftware.net] (Tamura et al., 2013). The phylogenetic tree was generated by bootstrap test Neighbour joining (NJ) with kimura two-parameter model (Kimura, 1980). The stability of internal nodes was assessed by bootstrap analysis with 1000 replicates.
Acknowledgement
The authors wish to acknowledge to Department of Biotechnology, Government of India for providing financial assistance under PG teaching HRD program.
References
Ahuja S. C., Panwar D.V.S., Ahuja U., and Gupta K.R., 1995, Basmati Rice - The Scented Pearl, CCS Haryana Agricultural University, Hissar, India, p.63
Ammiraju J., Lu F., and Sanyal A., 2008, Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell, 20: 3191-3209
http://dx.doi.org/10.1105/tpc.108.063727
Brar D.S., and Khush G.S., 1997, Alien introgression in rice. Plant Molecular Biology, 35: 35-47
http://dx.doi.org/10.1023/A:1005825519998
Cameron K.M., 2005, Leave it to the leaves: a molecular phylogenetic study of Malaxideae (Orchidaceae), American Journal of Botany, 92: 1025-1032
http://dx.doi.org/10.3732/ajb.92.6.1025
Clarck L. G., Zhang W., and Wendel J. F., 1995, A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Systematic Botany, 20: 436-460
http://dx.doi.org/10.2307/2419803
Clegg M.T., Gaut B. S., Learn G. H., and Morton B.R.,1994, Rates and patterns of chloroplast DNA evolution. Proceedings of the National Academy of Sciences, USA, 91: 6795-6801
http://dx.doi.org/10.1073/pnas.91.15.6795
Daniell H., Lee S.B., Grevich J., Saski C., Vargas T.Q., Guda C.B., Tomkins J., and Jansen R.K., 2006, Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes, Theoretical and Applied Genetics, 112: 1503-1518
http://dx.doi.org/10.1007/s00122-006-0254-x
Edwards A., Civitello A., Hammond H.A., and Casey C.T., 1991, DNA typing and genetic mapping with trimeric and tetrameric tendem repeats. American Journal of Human Genetics., 49: 746- 756
Goldman D.H., Freudenstein J.V., Kores P.J., Molvray M., Jarrell D.C., Whitten W.M., Cameron K.M., Jansen R.K., and Chase M.W., 2001, Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Systematic Botany, 26:670- 695
Hayashi K., and Kawano S., 2000, Molecular systematic of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data, Plant Species Biology, 15:73-93
http://dx.doi.org/10.1046/j.1442-1984.2000.00025.x
Hilu K.W., and Alice L.A., 1999, Evolutionary implications of matK indels in Poaceae, American Journal of Botany, 86:1735-1741
http://dx.doi.org/10.2307/2656671
Hilu K.W., Borsch T., Müller K., Soltis D.E., Soltis P.S., Savolainen V., Chase M.W., Powell M.P., Alice L.A., Evans R., Sauquet H., Neinhuis C., Slotta T.A.B., Jens G.R., Campbell C.S., and Chatrou L.W., 2003, Angiosperm phylogeny based on matK sequence information, American Journal of Botany, 90: 1758-1776
http://dx.doi.org/10.3732/ajb.90.12.1758
Hsiao C., Chatterton N.J., Asay K. H., and Jensen K.B., 1994, Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome, 37: 112-120
http://dx.doi.org/10.1139/g94-014
Jarrel D.C., and Clegg M.T.,1995, Systematic implications of the chloroplast-encoded matK gene on the tribe Vandeae (Orchidaceae), American Journal of Botany, 82: 137-145
Johnson L. A., and Soltis E., 1994, matK DNA sequences and phylogenetic reconstruction in Saxifragaceae sensustricto. Systematic Botany, 19: 143-156
http://dx.doi.org/10.2307/2419718
Johnson L. A.,and Soltis E., 1995, Phylogenetic inference in Saxifragaceae sensustricto and Gilia (Polemoniaceae) using matK sequences, Annals of the Mossouri Botanic Garden,82: 149-175
http://dx.doi.org/10.2307/2399875
Khush G. S., 2000, Taxonomy and origin of rice. In: Singh R. K., Singh U.S. and Khush G.S. (eds), Aromatic rices, Mohan Primlani for Oxford & IBH Publishing Co. Pvt. Ltd, New Delhi, India, PP. 5-13
Kimura M., 1980, A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16: 111-120
http://dx.doi.org/10.1007/BF01731581
Kores P.J., Weston P.H., Molvray M., and Chase M.W., 2000, Phylogenetic relationships within the Diurideae (Orchidaceae): inferences from plastid matK DNA sequences. In Wilson K.L. and Morrison D.A. (eds.), Monocots: Systematic and Evolution, CSIRO Publishing, Australia, 449-456
Kron K. A., Fuller R., Crayn D. M., Gadek P. A., and Quinn C. J., 1999, Phylogenetic relationships of epacrids and vaccinioids (Ericaceaes. l.) based on matK sequence data, Plant Systematic Evolution., 218:55-65
http://dx.doi.org/10.1007/BF01087034
Liang H., and Hilu K.W., 1996, Application of the matK gene sequences to grass systematic. Canadian Journal of Botany, 74: 125-134
http://dx.doi.org/10.1139/b96-017
Lu F., Ammiraju J.S.S., and Sanyal A., 2009, Comparative sequence analysis of MONOCULM1- orthologous regions in 14 Oryza genomes, Proceedings of the National Academy of Science, USA, 106: 2071-2076
http://dx.doi.org/10.1073/pnas.0812798106
Muller K.F., Borsch T., and Hilu K.W., 2006, Phylogenetic utility of rapidly evolving DNA at high taxonomical levels: contrasting matK, trnT-F and rbcL in basal angiosperms, Molecular Phylogenetics and Evolution, 41: 99-117
http://dx.doi.org/10.1016/j.ympev.2006.06.017
Olmstead R.G., and Palmer J.D., 1994, Chloroplast DNA systematics: a review of methods and data analysis, American Journal of Botany, 81: 1205-1224
http://dx.doi.org/10.2307/2445483
Sanger F., Nicklen S., and CoulsonA. R., 1977, DNA sequencing with chain terminating inhibitors, Proceedings of the National Academy of Science, USA. 74:5463-5467
http://dx.doi.org/10.1073/pnas.74.12.5463
Sastri D. C., HiluK. W., Appels R., Lagudah E. S., Playford J., and Baum B. 1992, An overview of evolution at the 5S RNA gene loci in plants. Plant Systematics and Evolution, 183: 169-181
http://dx.doi.org/10.1007/BF00940801
Soltis D.E., and Soltis P.S., 2004, Amborella not a “basal angiosperm”? Not so fast, American Journal of Botany, 91: 997-1000
http://dx.doi.org/10.3732/ajb.91.6.997
Steane D.A., 2005, Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae), DNA Research, 12: 215-220
http://dx.doi.org/10.1093/dnares/dsi006
Steele K.P., and Vilgalay R., 1994, Phylogenetic analyses of Polemoniaceae using nucleotide sequences of the Plastid gene matK. Systematic Botany, 19: 126-142
Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S., 2013, MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution, 30:2725-2729
http://dx.doi.org/10.1093/molbev/mst197
Turmel M., Otis C., and Lemieux C., 2006, The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants, Molecular Biology and Evolution, 23: 1324-1338
http://dx.doi.org/10.1093/molbev/msk018
. PDF(1150KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Rahul G. Patil
. Kundansingh R. Jadhao
. Kailash C. Samal
. Gyana R. Rout
Related articles
. Aromatic rice
. matK
. Molecular phylogeny
. Evolution
Tools
. Email to a friend
. Post a comment
.jpg)


