Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 5 doi: 10.5376/mgg.2024.15.0024
Received: 15 Aug., 2024 Accepted: 26 Sep., 2024 Published: 13 Oct., 2024
Zhou L., and Jiang L., 2024, High-throughput sequencing in maize: a gateway to precision breeding, Maize Genomics and Genetics, 15(5): 247-256 (doi: 10.5376/mgg.2024.15.0024)
High-throughput sequencing (HTS) technologies have revolutionized maize breeding by enabling precise genomic analysis and the identification of beneficial traits. This review explores the impact of HTS on maize, highlighting its role in discovering quantitative trait loci (QTL), genes, and alleles that contribute to crop improvement. The integration of multi-omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, has significantly advanced our understanding of maize's response to abiotic stresses. HTS has facilitated the assembly of complex genomes, identification of genetic variations, and the development of molecular markers for precision breeding. Despite challenges such as data analysis and cost, HTS remains a cornerstone in the pursuit of enhanced maize productivity and resilience.
1 Introduction
Maize (Zea mays) is one of the most significant staple crops globally, serving as a crucial source of food, animal feed, and industrial raw materials. Its importance is underscored by its extensive cultivation and the diverse applications it supports, from human consumption to biofuel production (Wang et al., 2022; Jafari et al., 2023). The crop's adaptability and high yield potential have made it a cornerstone of global food security and agricultural sustainability (Andorf et al., 2019; Wang et al., 2022). Historically, maize has undergone significant genetic and phenotypic transformations since its domestication from teosinte, which has contributed to its current status as a vital agricultural commodity (Liu et al., 2019).
The advent of high-throughput sequencing (HTS) technologies has revolutionized the field of plant genomics, providing unprecedented insights into the genetic makeup of crops like maize. These technologies enable the rapid sequencing of large genomes, facilitating the identification of genetic variations and the understanding of complex traits (Andorf et al., 2019). HTS has been instrumental in advancing maize breeding programs by allowing for the precise manipulation of genetic material, thereby accelerating the development of new varieties with desirable traits such as increased yield, stress resistance, and improved nutritional content. The integration of HTS with other biotechnological tools, such as CRISPR-Cas genome editing, has further enhanced the potential for precision breeding in maize (Dong et al., 2019; Wang et al., 2022).
This study provides a comprehensive overview of the impact of high-throughput sequencing on maize breeding, explores the historical context and current applications of HTS in maize genomics, highlighting key advancements and breakthroughs. Additionally, this study will discuss the integration of HTS with other genomic tools and its implications for future breeding strategies. By examining the technological progress and its practical applications, this study seeks to underscore the transformative potential of HTS in achieving precision breeding in maize, ultimately contributing to global food security and sustainable agriculture.
2 Fundamentals of High-Throughput Sequencing Technology
2.1 Principles and types of high-throughput sequencing
High-throughput sequencing (HTS) technologies have revolutionized the field of genomics by enabling the rapid sequencing of large amounts of DNA and RNA. The fundamental principle of HTS involves the parallel sequencing of millions of small DNA fragments, which are then computationally assembled to reconstruct the original sequence. This approach contrasts with traditional Sanger sequencing, which sequences DNA fragments one at a time. HTS technologies have significantly reduced the cost and time required for sequencing, making it feasible to sequence entire genomes, transcriptomes, and other complex genetic materials (Dalca and Brudno, 2010; Reon and Dutta, 2016; Lee, 2023).
Several HTS platforms are commonly used in genomic research, each with its unique advantages and limitations. Illumina sequencing, one of the most widely used platforms, is known for its high accuracy and throughput, making it suitable for a wide range of applications, including whole-genome sequencing and RNA sequencing. Pacific Biosciences (PacBio) sequencing offers long-read capabilities, which are beneficial for resolving complex genomic regions and structural variants. Oxford Nanopore Technologies (ONT) provides real-time sequencing and ultra-long reads, which are advantageous for de novo genome assembly and the detection of epigenetic modifications. Each of these platforms has contributed to the advancement of genomic research by providing diverse tools to address different scientific questions (Caspar et al., 2018; Pradhan et al., 2019; Lee, 2023).
2.2 The current status of maize genome sequencing
The sequencing of the maize genome has undergone significant advancements over the past decade. Initial efforts focused on generating a reference genome for maize, which provided a foundation for subsequent research. The first draft of the maize genome was published in 2009, marking a milestone in plant genomics. Since then, continuous improvements in sequencing technologies and bioinformatics tools have enabled more detailed and accurate assemblies of the maize genome. These advancements have facilitated the identification of genetic variations and the understanding of complex traits in maize, contributing to crop improvement and precision breeding efforts (Pérez-Losada et al., 2020; Farooqi et al., 2022).
Several reference genomes for maize have been completed, each providing valuable insights into the genetic architecture of this important crop. Notable projects include the B73 reference genome, which has been extensively used in maize research, and the more recent NAM (Nested Association Mapping) population genomes, which offer a broader representation of maize genetic diversity. These reference genomes have been instrumental in identifying quantitative trait loci (QTL), genes, and alleles associated with important agronomic traits. Research projects leveraging these reference genomes have advanced researchers understanding of maize biology and facilitated the development of improved maize varieties with enhanced yield, stress tolerance, and nutritional quality (Reon and Dutta, 2016; Farooqi et al., 2022).
2.3 Data processing and analysis
Processing HTS data involves several critical steps to ensure accurate and reliable results. The initial step is quality control, where raw sequencing reads are assessed and filtered to remove low-quality data. This is followed by read alignment, where the filtered reads are mapped to a reference genome or assembled de novo. Subsequent steps include variant calling, where genetic variants such as single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) are identified, and annotation, where the functional implications of these variants are determined. Each of these steps requires specialized bioinformatics tools and algorithms to handle the large volume of data generated by HTS (Dalca and Brudno, 2010; Altmann et al., 2012; Guo et al., 2017).
Bioinformatics tools play a crucial role in the analysis of maize HTS data, enabling researchers to extract meaningful insights from complex datasets. Tools for read alignment, such as BWA and Bowtie, are used to map sequencing reads to the maize reference genome. Variant calling tools, such as GATK and SAMtools, identify genetic variants, while annotation tools, such as ANNOVAR and SnpEff, predict the functional impact of these variants. Additionally, specialized software for transcriptome analysis, such as Cufflinks and DESeq, are used to study gene expression patterns. The integration of these tools allows researchers to uncover the genetic basis of important traits in maize and apply this knowledge to precision breeding programs (Altmann et al., 2012; Pradhan et al., 2019; Quijada et al., 2020).
3 Applications of High-Throughput Sequencing in Maize Precision Breeding
3.1 Genomic selection
High-throughput sequencing (HTS) has significantly enhanced the efficiency of genomic selection (GS) in maize breeding by enabling the rapid and accurate identification of genetic markers associated with desirable traits. The integration of HTS with GS allows breeders to predict the performance of breeding lines based on their genomic profiles, thus accelerating the breeding cycle and increasing genetic gains. For instance, the development of genotyping by target sequencing (GBTS) platforms has provided affordable and high-quality genotyping options, which are crucial for marker-assisted selection and genomic selection in maize (Guo et al., 2019). Additionally, HTS facilitates the collection of comprehensive genomic data, which can be used to improve the accuracy of genomic predictions and optimize breeding strategies (Cabrera-Bosquet et al., 2012; Wang and Zhang, 2024).
Several case studies have demonstrated the effectiveness of HTS in improving maize yield and resistance to various stresses through genomic selection. For example, the integration of genomic-enabled prediction and high-throughput phenotyping has been shown to enhance the prediction accuracy for grain yield in drought-stressed and heat-stressed environments, thereby aiding the development of climate-resilient maize varieties (Juliana et al., 2018). Another study highlighted the use of HTS in developing SNP marker panels, which were validated and used for genomic selection to improve traits such as yield and stress resistance in maize breeding programs (Guo et al., 2019).
3.2 Integration of genome editing and HTS
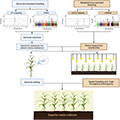
HTS plays a crucial role in detecting and validating genome editing events in maize. By providing detailed genomic information, HTS enables the precise identification of edits made by genome editing tools such as CRISPR/Cas9. This technology allows researchers to confirm the presence of intended modifications and assess any off-target effects, ensuring the accuracy and safety of genome editing applications (Figure 1) (Liu et al., 2020). The use of HTS in pre- and post-editing analysis ensures that only the desired genetic changes are retained, thereby streamlining the breeding process.
|
Figure 1 Pipeline of High-Throughput Genome-Editing Design (Adopted from Liu et al., 2020) Image caption: (A) Candidates selected from QTL fine mapping, genome-wide association mapping studies (GWAS), and comparative genomics; (B) Line-specific sgRNA filtering based on assembled pseudo-genome of the receptor line KN5585; (C) Different vector construction approaches of double sgRNA pool (DSP) and single sgRNA pool (SSP); (D) Measuring the coverage and uniformity during plasmid pool by deep-sequencing; (E) to (G) Transformation and assignment of targets to each T0 individual by barcode-based sequencing. (H) to (J) Identification of mutant sequences by Sanger sequencing; (K) and (L) Identification of mutant sequences by Capture-based deep-sequencing; (M) Measuring phenotype changes and identification of functional genes (Adopted from Liu et al., 2020) |
HTS is instrumental in verifying the effects of CRISPR/Cas9 and other genome editing techniques in maize. By sequencing the edited regions, researchers can determine the efficiency and specificity of the editing process. For instance, a study demonstrated the use of HTS to identify and characterize gene-editing events in maize, revealing the presence of homology-directed repair and other editing outcomes (Liu et al., 2020). This verification process is essential for understanding the functional impact of the edits and for optimizing genome editing protocols to achieve desired agronomic traits.
3.3 Mutant screening and gene mapping
HTS has revolutionized the establishment and screening of mutant libraries in maize. By enabling the rapid sequencing of large numbers of mutants, HTS allows for the comprehensive identification of genetic variations and their associated phenotypes. This high-throughput approach facilitates the discovery of novel genes and alleles that contribute to important agronomic traits, thereby accelerating the breeding process (Farooqi et al., 2022). The integration of HTS with mutant screening provides a powerful tool for functional genomics studies and the identification of key regulatory genes in maize.
The rapid identification and mapping of genes controlling important agronomic traits in maize have been greatly enhanced by HTS. This technology allows for the precise localization of quantitative trait loci (QTL) and the identification of candidate genes associated with traits such as yield, stress tolerance, and disease resistance. For example, HTS has been used to map genes related to abiotic stress tolerance in maize, providing valuable insights into the genetic basis of stress adaptation (Figure 2) (Farooqi et al., 2022). Additionally, the use of phased genotyping-by-sequencing has improved the analysis of genetic diversity and the identification of copy number variants, further aiding gene mapping efforts (Manching et al., 2017).
|
Figure 2 Types of abiotic stresses that affect yield productivity in maize (Adopted from Farooqi et al., 2022) |
4 Role of High-Throughput Sequencing in Maize Genetic Diversity Studies
4.1 Assessment of genetic diversity
High-throughput sequencing (HTS) technologies, such as genotyping-by-sequencing (GBS), have revolutionized the evaluation of genetic diversity in maize germplasm. For instance, the comprehensive genotyping of the USA national maize inbred seed bank utilized GBS to genotype 2 815 maize inbred accessions, resulting in the identification of 681 257 single-nucleotide polymorphism (SNP) markers across the genome. This extensive genotypic data revealed significant population stratification and moderate differentiation among major maize subpopulations, highlighting the utility of HTS in capturing the genetic diversity present in maize germplasm collections (Romay et al., 2013). Additionally, the use of inter-retrotransposon-amplified polymorphisms (IRAPs) has been effective in assessing genetic diversity, with studies showing high polymorphism rates and the ability to identify genetic similarities among maize lines (Ghonaim et al., 2020).
HTS data supports the conservation and utilization of maize germplasm by providing detailed genetic information that can guide breeding programs. For example, the development of high-resolution multiple-SNP arrays through genotyping by target sequencing (GBTS) has enabled the creation of marker panels that are powerful for genetic diversity detection and linkage disequilibrium decay analysis. These tools facilitate genome-wide association studies and the efficient management of genetic resources, ensuring that diverse germplasm is conserved and utilized effectively in breeding programs (Guo et al., 2019; Guo et al., 2021).
4.2 Population structure analysis and genetic linkage mapping
HTS data is instrumental in conducting population structure analysis in maize. The genotyping of diverse maize inbred lines using RNA-sequencing (RNA-seq) identified over 351 710 polymorphic loci, which were used to reveal tight clustering of distinct heterotic groups and exotic lines. This clustering provides insights into the genetic structure of maize populations, which is crucial for understanding the genetic basis of important traits and for designing effective breeding strategies (Hansey et al., 2012). Similarly, the phased genotyping-by-sequencing approach has enhanced the analysis of genetic diversity and revealed divergent copy number variants, further contributing to the understanding of population structure (Manching et al., 2017).
HTS technologies have enabled the construction of high-density genetic linkage maps, which are essential for quantitative trait loci (QTL) mapping of complex traits in maize. For instance, the GBS approach has been used to map roughly 200 000 sequence tags in maize recombinant inbred populations, providing a dense genetic map that supports the identification of QTLs associated with important agronomic traits (Elshire et al., 2011). These high-density maps are invaluable for dissecting the genetic architecture of complex traits and for facilitating marker-assisted selection in breeding programs.
4.3 Natural populations and domestication history studies
HTS has provided new insights into the domestication history and evolutionary processes of maize. By analyzing genetic diversity and population structure, researchers can trace the origins and spread of domesticated maize. For example, the comprehensive genotyping of maize germplasm has revealed patterns of genetic variation that reflect the domestication and subsequent breeding of maize, shedding light on the evolutionary processes that have shaped its current genetic makeup (Romay et al., 2013; Ghonaim et al., 2020).
HTS data has also been used to study gene flow between wild maize and domesticated varieties. The detailed genetic information obtained from HTS allows researchers to detect introgression events and to understand the impact of gene flow on the genetic diversity of domesticated maize. This information is crucial for maintaining genetic diversity and for the continued improvement of maize through breeding programs (Hansey et al., 2012; Romay et al., 2013).
5 Applications of High-Throughput Sequencing in Resistance Breeding
5.1 Identification of disease resistance genes
High-throughput sequencing (HTS) has significantly advanced the identification of disease resistance genes in maize. By enabling the comprehensive analysis of genetic variations, HTS facilitates the discovery of quantitative trait loci (QTL) and specific genes associated with disease resistance. This technology allows for the detailed mapping of resistance genes across the maize genome, providing a robust foundation for breeding programs aimed at enhancing disease resistance (Miedaner et al., 2020).
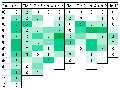
Several case studies have demonstrated the efficacy of HTS in disease resistance breeding. For instance, HTS has been employed to identify resistance genes against Northern corn leaf blight (NCLB) and Fusarium ear rot (FER) in maize. These studies have revealed that quantitative disease resistances are often controlled by multiple QTL scattered across the genome, which can be effectively targeted using HTS. Additionally, the integration of HTS with genomic selection has accelerated the breeding process by predicting the genomic estimated breeding values of untested genotypes, thereby enhancing the efficiency of resistance breeding programs (Figure 3) (Miedaner et al., 2020).
|
Figure 3 Number of QTLs for resistance to NCLB according to the literature; the intensity of color represents the number of QTLs found in the same bin (Adopted from Miedaner et al., 2020) |
5.2 Environmental stress resistance studies
HTS plays a crucial role in studying maize resistance to various environmental stresses such as drought and salinity. By enabling the comprehensive analysis of the maize genome, HTS helps identify key genes and regulatory networks involved in stress responses. This technology has facilitated the discovery of beneficial QTL and alleles that contribute to abiotic stress tolerance, thereby providing valuable insights for developing stress-resistant maize varieties (Farooqi et al., 2022).
HTS has been instrumental in analyzing gene expression and regulatory networks in maize under stress conditions. By employing transcriptomic approaches, researchers can identify differentially expressed genes and elucidate the underlying molecular mechanisms of stress tolerance. This information is critical for pinpointing specific genes that confer resistance to environmental stresses, which can then be targeted in breeding programs to develop more resilient maize varieties (Farooqi et al., 2022).
5.3 Development of molecular markers for insect resistance
HTS technology has revolutionized the development of molecular markers associated with insect resistance in maize. By enabling the high-throughput genotyping of large populations, HTS facilitates the identification of single nucleotide polymorphisms (SNPs) and other genetic markers linked to insect resistance traits. These markers can be used in marker-assisted selection (MAS) to accelerate the breeding of insect-resistant maize varieties (Guo et al., 2019).
A notable example of HTS application in insect resistance breeding is the development of SNP marker panels through genotyping by target sequencing (GBTS). This approach has been used to create affordable and high-quality marker panels that are highly consistent and reliable for MAS. The integration of these marker panels with other breeding platforms has greatly facilitated the molecular breeding activities, particularly in small- and medium-sized breeding programs. The use of HTS in developing these markers has proven to be cost-effective and efficient, making it a valuable tool for enhancing insect resistance in maize (Guo et al., 2019).
6 Future Prospects of High-Throughput Sequencing in Maize Breeding
6.1 Integration and utilization of multi-omics data
The integration of high-throughput sequencing (HTS) with various omics data, such as transcriptomics, epigenomics, proteomics, and metabolomics, holds significant promise for maize breeding. By combining these diverse data types, researchers can gain a comprehensive understanding of the genetic and molecular mechanisms underlying important agronomic traits. For instance, the integration of transcriptomic data with genomic information has been shown to improve the prediction of hybrid performance in maize, particularly for complex traits like dry matter yield (Westhues et al., 2017; Schrag et al., 2018). Additionally, multi-omics approaches have been successfully applied to identify novel biological markers for improving abiotic stress tolerance in maize (Yang et al., 2021; Farooqi et al., 2022).
The integration of multi-omics data can enhance the precision of breeding outcomes by providing a more detailed and holistic view of the genetic architecture of traits. This approach allows for the identification of key regulatory networks and interactions that are not captured by genomic data alone. For example, combining genomic, transcriptomic, and metabolomic data has been shown to improve the prediction of hybrid performance in maize, leading to more efficient selection of superior candidates (Schrag et al., 2018). Furthermore, the use of multi-omics data can help elucidate the molecular basis of phenotypic variations, thereby improving the accuracy of breeding value estimation (Yang et al., 2017; Yang et al., 2021; Zhou and Yan, 2024).
6.2 Advances and breakthroughs in HTS technology
Recent advancements in HTS technologies, such as single-cell sequencing, are poised to revolutionize maize breeding by providing unprecedented resolution and insights into cellular and molecular processes. Single-cell sequencing allows for the analysis of gene expression and genetic variation at the individual cell level, enabling the identification of rare cell types and the study of cellular heterogeneity. These advancements can lead to a better understanding of the genetic and epigenetic regulation of important traits, ultimately facilitating the development of more precise breeding strategies (Franzosa et al., 2015; Ritchie et al., 2015).
The potential applications of HTS in maize breeding are vast, ranging from the identification of beneficial quantitative trait loci (QTL) and genes to the development of precision breeding techniques. However, several challenges must be addressed to fully realize the potential of HTS. These include the need for advanced data integration and analysis methods to handle the large and complex datasets generated by HTS, as well as the development of cost-effective and scalable sequencing technologies (Ritchie et al., 2015; Yang et al., 2021; Farooqi et al., 2022). Additionally, the successful application of HTS in maize breeding will require overcoming technical and logistical challenges related to data management and sharing.
6.3 Challenges and opportunities in data management and sharing
The management and sharing of large-scale HTS data present significant challenges due to the sheer volume and complexity of the data. Effective data management strategies are essential to ensure the accessibility, reproducibility, and long-term storage of HTS data. This includes the development of standardized protocols for data collection, processing, and storage, as well as the implementation of robust data sharing platforms that facilitate collaboration among researchers (Franzosa et al., 2015; Ritchie et al., 2015). Addressing these challenges will be critical to maximizing the utility of HTS data in maize breeding.
Promoting international data sharing is crucial for driving collaborative research and accelerating progress in global maize breeding. By sharing HTS data and associated metadata, researchers can leverage collective knowledge and resources to address common challenges and develop innovative solutions. International data sharing initiatives can also help standardize methodologies and ensure the comparability of results across different studies. Collaborative efforts in data sharing have the potential to enhance the efficiency and effectiveness of maize breeding programs worldwide, ultimately contributing to global food security (Yang et al., 2021; Farooqi et al., 2022; Franzosa et al., 2022).
7 Concluding Remarks
High-throughput sequencing (HTS) technologies have significantly advanced maize breeding by enabling the identification of beneficial quantitative trait loci (QTL), genes, and alleles crucial for crop improvement. The integration of multi-omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, has been particularly effective in addressing abiotic stress tolerance in maize, leading to the discovery of novel biological markers. Additionally, the development of genotyping by target sequencing (GBTS) platforms has provided affordable and high-quality genotyping options, facilitating marker-assisted breeding and enhancing the efficiency of molecular breeding activities. HTS has also revolutionized plant molecular breeding by offering ultra-low cost per base of sequencing and high data output, enabling comprehensive studies in crop genetics and genomics.
HTS is poised to continue driving advancements in maize breeding technologies. The ongoing development of more sophisticated and cost-effective sequencing platforms will further enhance the precision and efficiency of breeding programs. The integration of HTS with other breeding platforms and open-source networks is expected to democratize access to advanced breeding tools, particularly benefiting small- and medium-sized enterprises and developing countries. The prospects for widespread application of precision breeding in agriculture are promising, with HTS enabling more accurate selection of desirable traits and faster development of stress-resistant and high-yielding maize varieties.
The importance of continued technological innovation and research in HTS cannot be overstated. As HTS technologies evolve, they will unlock new possibilities for understanding and manipulating the maize genome, leading to more resilient and productive crops. The future of maize breeding science holds great promise, with HTS at the forefront of this transformation. Continued investment in HTS research and development will be crucial in meeting the global challenges of food security and climate change, ensuring sustainable agricultural practices and improved crop yields.
Acknowledgments
We would like to thank Mr Xuan for providing the plant samples used in this study.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Altmann A., Weber P., Bader D., Preuss M., Binder E., and Müller-Myhsok B., 2012, A beginners guide to SNP calling from high-throughput DNA-sequencing data, Human Genetics, 131: 1541-1554.
https://doi.org/10.1007/s00439-012-1213-z
PMID: 22886560
Andorf C., Beavis W., Hufford M., Smith S., Suza W., Wang K., Woodhouse M., Yu J.M., and Lübberstedt T., 2019, Technological advances in maize breeding: past, present and future, Theoretical and Applied Genetics, 132: 817-849.
https://doi.org/10.1007/s00122-019-03306-3
PMID: 30798332
Cabrera-Bosquet L., Crossa J., von Zitzewitz J., Serret M.D., and Araus J.L., 2012, High-throughput phenotyping and genomic selection: the frontiers of crop breeding converge, Journal of Integrative Plant Biology, 54(5): 312-320.
https://doi.org/10.1111/j.1744-7909.2012.01116.x
Caspar S.M., Dubacher N., Kopps A.M., Meienberg J., Henggeler C., and Matyas G., 2018, Clinical sequencing: from raw data to diagnosis with lifetime value, Clinical Genetics, 93(3); 508-519.
https://doi.org/10.1111/cge.13190
Dalca A.V., and Brudno M., 2010, Genome variation discovery with high-throughput sequencing data, Briefings in Bioinformatics, 11(1): 3-14
https://doi.org/10.1093/bib/bbp058
Dong L., Qi X.T., Zhu J.J., Liu C.L., Zhang X., Cheng B.J., Mao L., and Xie C.X., 2019, Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing, Plant Biotechnology Journal, 17(10): 1853-1855.
https://doi.org/10.1111/pbi.13144
Elshire R.J., Glaubitz J.C., Sun Q., Poland J.A., Kawamoto K., Buckler E.S., and Mitchell S.E., 2011, A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species, PLoS One, 6(5): e19379.
https://doi.org/10.1371/journal.pone.0019379
PMID: 21573248 PMCID: PMC3087801
Farooqi M.Q.U., Nawaz G., Wani S.H., Choudhary J.R., Rana M., Sah R.P., Afzal M., Zahra Z., Ganie S.A., Razzaq A., Reyes V.P., Mahmoud E.A., Elansary H.O., El-Abedin T., and Siddique K., 2022, Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.), Frontiers in Plant Science, 13: 965878.
https://doi.org/10.3389/fpls.2022.965878
PMID: 36212378 PMCID: PMC9538355
Franzosa E.A., Hsu T., Sirota-Madi A., Shafquat A., Abu-Ali G., Morgan X.C., and Huttenhower C., 2015, Sequencing and beyond: integrating molecular 'omics' for microbial community profiling, Nature Reviews Microbiology, 13(6): 360-372.
https://doi.org/10.1038/nrmicro3451
PMID: 25915636 PMCID: PMC4800835
Ghonaim M., Kalendar R., Barakat H., Elsherif N., Ashry N., and Schulman A.H., 2020, High-throughput retrotransposon-based genetic diversity of maize germplasm assessment and analysis, Molecular Biology Reports, 47(3): 1589-1603.
https://doi.org/10.1007/s11033-020-05246-4
Guo Y., Han L., and Sheng Q.H,, 2017, Recent advances in high throughput sequencing analysis, International Journal of Genomics, 2017: 2454780.
https://doi.org/10.1155/2017/2454780
PMID: 28706940 PMCID: PMC5494582
Guo Z.F., Wang H.W., Tao J.J., Ren Y.H., Xu C., Wu K.S., Zou C., Zhang J.N,, and Xu Y.B., 2019, Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize, Molecular Breeding, 39: 1-12.
https://doi.org/10.1007/s11032-019-0940-4
Guo Z.F., Yang Q.N., Huang F.F., Zheng H.J., Sang Z.Q., Xu Y.F., Zhang C., Wu K.S., Tao J.J., Prasanna B.M., Olsen M.S., Wang Y.B., Zhang J.A., and Xu Y.B., 2021, Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip, Plant Communications, 2(6): 100230.
https://doi.org/10.1016/j.xplc.2021.100230
PMID: 34778746 PMCID: PMC8577115
Hansey C.N., Vaillancourt B., Sekhon R.S., de León N., Kaeppler S.M., and Buell C.R., 2012, Maize (Zea mays L.) genome diversity as revealed by RNA-Sequencing, PLoS One, 7(3): e33071.
https://doi.org/10.1371/journal.pone.0033071
PMID: 22438891 PMCID: PMC3306378
Jafari F., Wang B.B., Wang H.Y., and Zou J.J., 2023, Breeding maize of ideal plant architecture for high-density planting tolerance through modulating shade avoidance response and beyond, Journal of Integrative Plant Biology, 66(5): 849-864.
https://doi.org/10.1111/jipb.13603
Juliana P., Montesinos-López O.A., Crossa J., Mondal S., Pérez L.G., Poland J., Huerta-Espino J., Crespo-Herrera L., Govindan V., Dreisigacker S., Shrestha S., Pérez-Rodríguez P., Espinosa F.P., and Singh R., 2018, Integrating genomic-enabled prediction and high-throughput phenotyping in breeding for climate-resilient bread wheat, Theoretical and Applied Genetics, 132(1): 177-194.
https://doi.org/10.1007/s00122-018-3206-3
PMID: 30341493 PMCID: PMC6320358
Lee J.Y., 2023, The principles and applications of high-throughput sequencing technologies, Development and Reproduction, 27(1): 9-24.
https://doi.org/10.12717/DR.2023.27.1.9
PMID: 38075439 PMCID: PMC10703097
Liu H.J., Jian L.M., Xu J.T., Zhang Q.H., Zhang M.L., Jin M.L., Peng Y., Yan J.L., Han B.Z., Liu J., Gao F., Liu X.G., Huang L., Wei W.J., Ding Y.X., Yang X.F., Li Z.X., Zhang M.L., Sun J.M., Bai M.J., Song W.H., Chen H.M., Sun X.A., Li W.Q., Lu Y.M., Liu Y., Zhao J.R., Qian Y.W., Jackson D., Fernie A.R., and Yan J.B., 2020, High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize, Plant Cell, 32(5): 1397-1413.
https://doi.org/10.1105/tpc.19.00934
PMID: 32102844 PMCID: PMC7203946
Liu J., Fernie A.R., and Yan J.B., 2019, The past, present, and future of maize improvement: domestication, genomics, and functional genomic routes toward crop enhancement, Plant Communications, 1(1): 100010.
https://doi.org/10.1016/j.xplc.2019.100010
PMID: 33404535 PMCID: PMC7747985
Manching H., Sengupta S., Hopper K.R., Polson S.W., Ji Y., and Wisser R.J., 2017, Phased genotyping-by-sequencing enhances analysis of genetic diversity and reveals divergent copy number variants in maize, G3: Genes|Genomes|Genetics, 7(7): 2161-2170.
https://doi.org/10.1534/g3.117.042036
Miedaner T., Boeven A.L., Gaikpa D.S., Kistner M.B., and Grote C.P., 2020, Genomics-assisted breeding for quantitative disease resistances in small-grain cereals and maize, International Journal of Molecular Sciences, 21(24): 9717.
https://doi.org/10.3390/ijms21249717
PMID: 33352763 PMCID: PMC7766114
Pérez-Losada M., Arenas M., Galán J.C., Bracho M.A., Hillung J., García-González N., and González-Candelas F., 2020, High-throughput sequencing (HTS) for the analysis of viral populations, Infection, Genetics and Evolution, 80: 104208.
https://doi.org/10.1016/j.meegid.2020.104208
Pradhan D., Kumar A., Singh H., and Agrawal U., 2019, High-throughput sequencing, Data Processing Handbook for Complex Biological Data Sources, 2019: 39-52.
https://doi.org/10.1016/B978-0-12-816548-5.00004-6
Quijada N.M., Hernández M., and Rodríguez-Lázaro D., 2020, High-throughput sequencing and food microbiology, Advances in Food and Nutrition Research, 91: 275-300.
https://doi.org/10.1016/bs.afnr.2019.10.003
Reon B.J., and Dutta A., 2016, Biological processes discovered by high-throughput sequencing, The American Journal of Pathology, 186(4): 722-732.
https://doi.org/10.1016/j.ajpath.2015.10.033
Ritchie M.D., Holzinger E.R., Li R., Pendergrass S.A., and Kim D., 2015, Methods of integrating data to uncover genotype–phenotype interactions, Nature Reviews Genetics, 16(2): 85-97.
https://doi.org/10.1038/nrg3868
PMID: 25582081
Romay M.C., Millard M.J., Glaubitz J.C., Peiffer J.A., Swarts K.L., Casstevens T.M., Elshire R.J., Acharya C.B., Mitchell S.E., Flint-Garcia S.A., McMullen M.D., Holland J.B., Buckler E.S., and Gardner C.A., 2013, Comprehensive genotyping of the USA national maize inbred seed bank, Genome Biology, 14(6): R55.
https://doi.org/10.1186/gb-2013-14-6-r55
PMID: 23759205 PMCID: PMC3707059
Schrag T.A., Westhues, M., Schipprack, W., Seifert, F., Thiemann, A., Scholten, S., and Melchinger, A., 2018, Beyond genomic prediction: combining different types of omics data can improve prediction of hybrid performance in maize, Genetics, 208(4): 1373-1385.
https://doi.org/10.1534/genetics.117.300374
PMID: 29363551 PMCID: PMC5887136
Wang Y.H., Tang Q.L., Pu L., Zhang H.W., and Li X.H., 2022, CRISPR-Cas technology opens a new era for the creation of novel maize germplasms, Frontiers in Plant Science, 13: 1049803.
https://doi.org/10.3389/fpls.2022.1049803
PMID: 36589095 PMCID: PMC9800880
Wang Y.F., and Zhang L.M., 2024, Gene-driven future: breakthroughs and applications of marker-assisted selection in tree breeding, Molecular Plant Breeding, 15(3): 132-143.
https://doi.org/10.5376/mpb.2024.15.0014
Westhues M., Schrag T.A., Heuer C., Thaller G., Utz H.F., Schipprack W., Thiemann A., Seifert F., Ehret A., Schlereth A., Stitt M., Nikoloski Z., Willmitzer L., Schön C.C., Scholten S., and Melchinger A., 2017, Omics-based hybrid prediction in maize, Theoretical and Applied Genetics, 130(9): 1927-1939.
https://doi.org/10.1007/s00122-017-2934-0
Yang Y.D., Saand M.A., Huang L.Y., Abdelaal W.B., Zhang J., Wu Y., Li J., Sirohi M.H., and Wang F.Y., 2021, Applications of multi-omics technologies for crop improvement, Frontiers in Plant Science, 12: 563953.
https://doi.org/10.3389/fpls.2021.563953
PMID: 34539683 PMCID: PMC8446515
Yang Y.R., Zhou R., and Kui L., 2017, Future livestock breeding: precision breeding based on multi-omics information and population personalization, Journal of Integrative Agriculture, 16(12): 2784-2791.
https://doi.org/10.1016/S2095-3119(17)61780-5
Zhou J.Y., and Yan S.D., 2024, A comprehensive review of corn ethanol fuel production: from agricultural cultivation to energy application, Journal of Energy Bioscience, 15(3): 208-220.
https://doi.org/10.5376/jeb.2024.15.0020

. PDF(996KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Lan Zhou
. Long Jiang
Related articles
. High-throughput sequencing (HTS)
. Maize breeding
. Multi-omics
. Quantitative trait loci (QTL)
. Abiotic stress tolerance
Tools
. Email to a friend
. Post a comment