 Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 4 doi: 10.5376/mgg.2024.15.0020
Received: 29 Jun., 2024 Accepted: 05 Aug., 2024 Published: 30 Aug., 2024
Yang X.J., and Song B.X., 2024, Advancements in maize genomic tools for enhanced crop breeding, Maize Genomics and Genetics, 15(4): 204-217 (doi: 10.5376/mgg.2024.15.0020)
The rapid advancements in maize genomic tools have significantly enhanced crop breeding efforts, addressing the urgent need for improved yield, resilience, and adaptability in the face of global challenges such as climate change and food security. This review highlights the historical development and current state of maize transformation technologies, including CRISPR/Cas9-mediated genome editing, which has revolutionized the efficiency and scope of genetic modifications in maize. The integration of high-throughput phenotyping and genomic selection has further accelerated the breeding cycle, enabling the precise selection of superior genotypes. Additionally, innovative approaches such as genomic design breeding and multiplex genome editing strategies have been proposed to optimize genetic gains and improve complex traits like drought tolerance and yield. These advancements underscore the critical role of genomics in modern maize breeding, paving the way for sustainable agricultural practices and enhanced global food security.
1 Introduction
Maize (Zea mays) is a cornerstone of global agriculture, serving as a vital food source, animal feed, and industrial raw material. It is one of the three major grain crops worldwide, alongside rice and wheat, and plays a crucial role in the diets of billions of people, particularly in developing regions such as sub-Saharan Africa, Southeast Asia, and Latin America (Nuss and Tanumihardjo, 2010). The crop's versatility extends beyond nutrition, contributing significantly to the global agricultural economy and the livelihoods of millions of farmers (Agarwal et al., 2018). The demand for maize is projected to increase substantially, driven by its use in food, feed, and bioenergy production, necessitating a 2% annual increase in global production to meet future needs (Ortiz et al., 2010).
The advent of genomic tools has revolutionized agricultural practices, enabling more precise, efficient, and rapid breeding of crops. Traditional breeding methods, while effective, are time-consuming and often limited by the genetic diversity available within elite breeding pools (Ortiz et al., 2010). Modern genomic tools, such as CRISPR/Cas9, marker-assisted selection, and genomic prediction, have opened new avenues for crop improvement by allowing targeted genetic modifications and the acceleration of breeding cycles (Agarwal et al., 2018; Thudi et al., 2020; Muntean et al., 2022). These tools facilitate the development of high-performance hybrids with enhanced traits such as increased yield, nutritional quality, and resistance to biotic and abiotic stresses (Wan et al., 2019; Muntean et al., 2022; Zhang et al., 2023). The integration of bioinformatics and advanced genomic resources further supports the identification and utilization of beneficial genetic variations, thereby enhancing the overall efficiency of breeding programs (Palacios-Rojas et al., 2020; Muntean et al., 2022).
This study aims to provide a comprehensive overview of the advancements in maize genomic tools and their applications in crop breeding. It will cover the historical context of maize domestication and breeding, highlight the key genomic technologies currently in use, and discuss their impact on maize improvement. The article will also explore future directions in maize genomics, emphasizing the potential of these tools to address the challenges posed by climate change and the growing global demand for food. By synthesizing the latest research and developments, this study seeks to underscore the critical role of genomic tools in enhancing maize breeding and ensuring food security in the face of evolving agricultural challenges.
2 Development of Maize Genomic Resources
2.1 Sequencing of the maize genome: milestones and progress
The sequencing of the maize genome has undergone significant advancements over the past decade, driven by the development of new sequencing technologies and methodologies. One of the most notable milestones was the assembly and annotation of the maize genome using Single Molecule Real-Time (SMRT) sequencing and high-resolution optical mapping. This approach resulted in a 52-fold increase in contig length compared to previous reference genomes, with substantial improvements in the assembly of intergenic spaces and centromeres. The characterization of the repetitive portion of the genome revealed over 130 000 intact transposable elements, providing insights into transposable element lineage expansions unique to maize. Additionally, gene annotations were updated using 111 000 full-length transcripts obtained by SMRT sequencing (Jiao et al., 2017).
Another significant contribution to maize genomics was the draft genome sequence of the elite maize line HuangZaoSi (HZS). This genome sequence provided insights into genomic variation and the improvement history of maize. The study revealed that more than 60% of identified selective sweeps were clustered in identity-by-descent conserved regions, and yield-related genes/QTLs were enriched in HZS characteristic selected regions. This research expanded our understanding of the breadth of genomic variation and the historical improvement of maize (Li et al., 2019).
2.2 Public databases and genomic repositories
Public databases and genomic repositories play a crucial role in the dissemination and utilization of maize genomic resources. The USA National Plant Germplasm System, for instance, has utilized genotyping-by-sequencing (GBS) to genotype 2 815 maize inbred accessions. This effort produced 681 257 single-nucleotide polymorphism (SNP) markers distributed across the entire genome, enabling the detection of rare alleles with high confidence. The genotypic information from this publicly available panel allows researchers to explore genetic diversity and perform genome-wide association studies, thereby addressing challenges in sustainable agriculture (Romay et al., 2013).
Genotyping-by-sequencing (GBS) has emerged as a powerful tool for marker-assisted selection (MAS) in plant breeding. GBS combines molecular marker discovery and genotyping, making it a cost-effective technique for large-scale plant breeding programs. It has been successfully used in genome-wide association studies (GWAS), genomic diversity studies, genetic linkage analysis, and molecular marker discovery, significantly accelerating the breeding process (Bevan et al., 2017).
2.3 Comparative genomics and evolutionary insights
Comparative genomics has provided valuable evolutionary insights into maize and its relatives. The detailed characterization of plant genomes and genetic diversity has been instrumental in identifying a wide spectrum of genetic variation and associating it with diverse agronomic phenotypes. Advances in genome sequencing and assembly have enabled researchers to access the large and complex genomes of crops and their wild relatives, facilitating the identification of genetic diversity and its association with agronomic traits (Figure 1) (Andorf et al., 2019).
 Figure 1 Timeline of sequencing technologies, major genomes and maize genomics (Adopted from Andorf et al., 2019) Image caption: The figure shows three timelines. The first timeline list the release dates of major sequencing technologies focusing on the early first-generation technologies (1950–1990) and the next-generation sequencing technologies (2000s). The second timeline shows the release dates of four major genomes (yeast, arabidopsis, human and rice) and the first reported pan-genome (bacteria). The third timeline shows the release dates of maize genomes and major genotype datasets (Adopted from Andorf et al., 2019) |
The comparative analysis of maize genomes has also revealed the prevalence of deletions in regions of low gene density and maize lineage-specific genes. This information is crucial for understanding the evolutionary history of maize and its adaptation to different environments (Jiao et al., 2017). Furthermore, the proteome clustering of six completed maize genomes identified 638 proteins falling into 264 HZS-specific gene families, with the majority of contributions from tandem duplication events. This study provided novel insights into the genomic variation and improvement history of maize, highlighting the importance of comparative genomics in crop improvement (Li et al., 2019).
In summary, the development of maize genomic resources has been marked by significant milestones in genome sequencing, the establishment of public databases and genomic repositories, and the application of comparative genomics. These advancements have provided a solid foundation for enhanced crop breeding, enabling researchers to explore genetic diversity, understand evolutionary processes, and develop improved maize varieties for sustainable agriculture.
3 Genomic Selection and Marker-Assisted Breeding
3.1 Principles of genomic selection in maize breeding
Genomic selection (GS) is a revolutionary approach in plant breeding that leverages genome-wide marker data to predict the breeding values of individuals. Unlike traditional marker-assisted selection (MAS), which focuses on a few markers associated with specific traits, GS uses all available marker data to estimate the genetic potential of breeding candidates. This comprehensive approach allows for the selection of superior genotypes with greater accuracy and efficiency, thereby accelerating the breeding cycle and enhancing genetic gains (Crossa et al., 2017; Rice and Lipka, 2021; Budhlakoti et al., 2022).
The principle of GS is rooted in the use of genomic-estimated breeding values (GEBVs), which are derived from statistical models that incorporate genome-wide marker information. These models predict the performance of untested genotypes based on their genetic makeup, enabling breeders to make informed decisions without extensive phenotypic evaluations. This is particularly advantageous for complex traits controlled by multiple genes with small effects, where traditional MAS falls short (Jannink et al., 2010; Budhlakoti et al., 2022).
GS has shown significant promise in maize breeding, where it has been applied to develop superior inbreds and hybrids. The integration of high-throughput phenotypic, genotypic, and other -omic data has further refined GS models, allowing for the encapsulation of non-additive genetic effects, genotype-by-environment interactions, and multiple levels of the biological hierarchy. These advancements have led to more accurate predictions of breeding values and, consequently, more efficient breeding programs (Crossa et al., 2017; Rice and Lipka, 2021).
3.2 Development and application of molecular markers
Molecular markers are essential tools in modern plant breeding, providing a means to track genetic variations associated with desirable traits. The development of various types of molecular markers, such as single nucleotide polymorphisms (SNPs), has revolutionized the field. These markers are used in MAS to assist phenotypic selections, thereby improving the accuracy and efficiency of breeding programs (Eathington et al., 2007; He et al., 2014; Hasan et al., 2021).
The advent of next-generation sequencing (NGS) technologies has further expanded the utility of molecular markers. Techniques like genotyping-by-sequencing (GBS) have enabled the simultaneous discovery and genotyping of SNPs in large crop genomes, such as maize. GBS involves the digestion of genomic DNA with restriction enzymes, followed by the ligation of barcode adapters, PCR amplification, and sequencing. This high-throughput approach has been successfully used in genome-wide association studies (GWAS), genetic diversity studies, and genomic selection, making it a cost-effective and powerful tool for large-scale plant breeding programs (He et al., 2014).
In addition to SNPs, other molecular markers such as quantitative trait loci (QTL) have been extensively mapped for various plant species. These markers have been used in MAS to improve traits like drought tolerance, disease resistance, and yield. The integration of molecular markers into breeding programs has significantly shortened the time required to develop new crop varieties, thereby enhancing the overall productivity and sustainability of agricultural systems (Varshney et al., 2013; Hasan et al., 2021).
3.3 Integration of genomic selection in breeding programs
The integration of GS into breeding programs requires a strategic approach to optimize its benefits. Traditional breeding programs need to be restructured to incorporate GS effectively. This involves reorganizing field designs, training populations, and increasing the number of lines evaluated. Leveraging large amounts of genomic and phenotypic data collected across different growing seasons and environments is crucial to increase heritability estimates, selection intensity, and selection accuracy (Merrick et al., 2022).
One of the key advantages of GS is its ability to improve selection accuracy while minimizing the need for extensive phenotyping. This is particularly beneficial for traits that are difficult or expensive to measure. By using GEBVs, breeders can select the best candidates for the next breeding cycle more efficiently, thereby reducing the overall time and cost associated with developing new varieties (Jannink et al., 2010; Budhlakoti et al., 2022).
GS has been successfully integrated into maize breeding programs, where it has been used to enhance traits such as yield, stress tolerance, and disease resistance. The use of advanced statistical models and high-throughput phenotyping techniques has further improved the accuracy of GS predictions. For instance, hyperspectral imaging technology can be combined with GS to provide detailed phenotypic data, which can be used to refine prediction models and enhance breeding outcomes (Crossa et al., 2017; Budhlakoti et al., 2022).
In conclusion, the integration of GS and molecular markers into maize breeding programs represents a significant advancement in crop improvement. By leveraging genome-wide marker data and advanced statistical models, breeders can achieve faster genetic gains and develop superior crop varieties with enhanced traits. The continued refinement and application of these genomic tools will play a crucial role in meeting the growing demands for food security and sustainable agriculture.
4 CRISPR and Genome Editing Techniques in Maize
4.1 Overview of CRISPR technology
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology, coupled with CRISPR-associated proteins (Cas), has revolutionized the field of genome editing. Initially discovered as a bacterial immune mechanism, CRISPR/Cas systems have been adapted for precise genome editing in various organisms, including plants and animals. The CRISPR/Cas9 system, in particular, has gained widespread use due to its simplicity, efficiency, and versatility (Figure 2) (Manghwar et al., 2019; Li et al., 2021; Wang and Doudna, 2023).
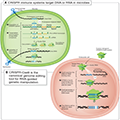 Figure 2 CRISPR-based adaptive immunity provides programmable genome editing tools (Adopted from Wang and Doudna, 2023) Image caption: (A) CRISPR immune systems target DNA or RNA in microbes (illustration depicts DNA targeting). Three steps to immunity include: (i) acquisition of CRISPR spacer sequence matching an infectious agent; (ii) transcription and formation of Cas-RNA complexes; (iii) seek-and-destroy surveillance mechanisms. (B) CRISPR-Cas9 is the canonical genome editing tool for RNA-guided genetic manipulation. Cas9 searches for target sites in a genome by engaging with PAM sequences, forming an R-loop with complementary DNA, generating a double-strand DNA (dsDNA) break, and finally releasing DNA for repair (Adopted from Wang and Doudna, 2023) |
CRISPR technology functions by utilizing a guide RNA (gRNA) to direct the Cas9 nuclease to a specific DNA sequence, where it introduces a double-strand break. This break can then be repaired by the cell's natural repair mechanisms, either through non-homologous end joining (NHEJ) or homology-directed repair (HDR), leading to targeted genetic modifications (Ran et al., 2013; Bortesi and Fischer, 2015). Recent advancements have expanded the CRISPR toolbox to include base editors, prime editors, and CRISPR interference (CRISPRi) and activation (CRISPRa) systems, which allow for more precise and varied genetic modifications (Kumlehn et al., 2018; Monsur et al., 2020; Li et al., 2021).
4.2 Applications of CRISPR in maize trait improvement
CRISPR technology has been instrumental in advancing maize breeding by enabling the precise modification of genes associated with important agronomic traits. Researchers have successfully used CRISPR/Cas9 to enhance traits such as yield, stress tolerance, and nutritional content in maize (Kumlehn et al., 2018; Li et al., 2021; Wang and Doudna, 2023).
One notable application is the improvement of maize's resistance to biotic and abiotic stresses. By targeting specific genes involved in stress response pathways, scientists have developed maize varieties with enhanced tolerance to drought, pests, and diseases. For example, CRISPR/Cas9 has been used to knock out genes that negatively regulate stress responses, thereby increasing the plant's resilience (Kumlehn et al., 2018; Li et al., 2021).
Additionally, CRISPR has been employed to improve the nutritional quality of maize. By editing genes involved in the biosynthesis of essential nutrients, researchers have created maize varieties with increased levels of vitamins, minerals, and other beneficial compounds. This has significant implications for addressing malnutrition and improving food security (Monsur et al., 2020; Wang and Doudna, 2023).
CRISPR technology has also facilitated the development of maize varieties with improved yield and growth characteristics. By targeting genes that regulate plant architecture, flowering time, and grain development, scientists have been able to create maize plants that produce higher yields and are better suited to different growing conditions (Kumlehn et al., 2018; Li et al., 2021).
4.3 Ethical considerations and regulatory framework
The rapid advancement of CRISPR technology has raised important ethical and regulatory considerations. One major concern is the potential for off-target effects, where unintended genetic modifications occur. While advancements in CRISPR technology have improved its precision, the risk of off-target effects remains, necessitating thorough evaluation and validation of edited plants (Bortesi and Fischer, 2015; Moon et al., 2019; Li et al., 2021).
Ethical considerations also extend to the broader implications of genome editing in agriculture. The use of CRISPR to create genetically modified organisms (GMOs) has sparked debates about food safety, environmental impact, and the socio-economic consequences for farmers and consumers. Public perception and acceptance of CRISPR-edited crops are critical factors that influence regulatory policies and market adoption (Bortesi and Fischer, 2015; Wang and Doudna, 2023).
Regulatory frameworks for CRISPR-edited crops vary globally. In some regions, such as the United States, CRISPR-edited crops are regulated similarly to traditional GMOs, with rigorous safety assessments and approval processes. In contrast, other regions, like the European Union, have stricter regulations that may limit the commercialization of CRISPR-edited crops. Harmonizing these regulatory approaches is essential to facilitate the global adoption of CRISPR technology in agriculture (Bortesi and Fischer, 2015; Moon et al., 2019; Wang and Doudna, 2023).
In conclusion, CRISPR technology has significantly advanced maize breeding by enabling precise genetic modifications that enhance important agronomic traits. However, ethical considerations and regulatory frameworks must be carefully navigated to ensure the safe and responsible use of this powerful technology. Continued research and dialogue among scientists, policymakers, and the public are crucial for realizing the full potential of CRISPR in agriculture.
5 Transcriptomics and Gene Expression Profiling
5.1 Advances in transcriptome sequencing technologies
The advent of advanced transcriptome sequencing technologies has revolutionized the field of maize genomics, providing unprecedented insights into gene expression and regulation. Single Molecule Real-Time (SMRT) sequencing, for instance, has significantly improved the assembly and annotation of the maize genome. This technology has enabled the identification of over 130 000 intact transposable elements and the generation of 111 000 full-length transcripts, which are crucial for understanding the functional and regulatory elements within the maize genome (Jiao et al., 2017). These advancements have facilitated the characterization of complex repeat regions and intergenic spaces, which were previously challenging to assemble accurately.
Moreover, large-scale transcriptome profiling across various tissues and developmental stages has provided a comprehensive view of gene expression dynamics in maize. For example, transcriptome profiling of 23 different tissues or developmental stages in maize inbreds B73 and Mo17, as well as their F1 hybrid, revealed extensive differential expression and regulatory variation. This study highlighted the dynamic nature of gene expression and the prevalence of tissue-specific regulatory mechanisms, which are essential for understanding phenotypic variation and heterosis in maize (Figure 3) (Zhou et al., 2019).
 Figure 3 Analysis of Biased Allelic Expression Patterns and Regulatory Variation Classification across Tissues (Adopted from Zhou et al., 2019) Image caption: (A and B) A scatterplot shows the parental B73 allele ratio (x axis, CPMB/(CPMB/CPMM)) and hybrid B73 allele ratio (y axis) for DE genes (A) and non-DE genes (B) in maize root tissue. The colors represent different regulatory variation classifications determined for each gene (see Methods) (C) The pie chart (left) shows the proportion of all DE genes (between the two parents) that were assigned to different regulatory mechanisms across all 20 non-endosperm tissues; The plots on the right show the enrichment or depletion (as fold change relative to background proportion from the left pie chart) for subsets of genes for each regulatory variation classification; The subsets of genes include different levels of fold change in expression (DE2–4, DE4–8, DE8+, and SPE), different additivity patterns (BLP, LP, MP, HP, and AHP), and genes that were characterized by a previous eQTL study (in shoot apex meristem, Li et al., 2013) to be regulated by only cis-eQTL, only trans-eQTL, or both cis-eQTL and trans-eQTL. For each subset of genes the proportion of each regulatory classification was compared with background proportion (left pie chart) with the ratio plotted as bars along the x axis. P values were determined using a hypergeometric test (lower.tail = FALSE for enrichment and lower.tail=TRUE for depletion) and labeled with asterisks (*P<0.01 or **P<0.001); (D and E) For genes that are DE and have allele-specific expression data in at least five tissues, we assessed the consistency of regulation variation classifications; In (D) the subset of genes that are classified into each pattern in at least one tissue was used to assess the proportion of tissues that show this pattern. In (E) all genes classified into a specific pattern in at least one tissue were used to assess whether that classification was tissue specific (showing that pattern in less than 20% of DE tissues), intermediate frequency (20%–80% DE tissues), or constitutive (more than 80% of DE tissues) (Adopted from Zhou et al., 2019) |
5.2 Functional genomics and gene expression studies
Functional genomics studies have been instrumental in identifying key regulatory genes and understanding their roles in maize development and agronomic traits. Gene expression and eQTL analyses have uncovered natural variations underlying important agronomic traits, such as plant architecture, flowering time, and yield-related traits. For instance, a study on 137 maize elite inbred lines identified several candidate genes, including ZmARF16, ZmARF34, and ZmTCP40, that regulate various plant architecture- and yield-related traits. These findings demonstrate the effectiveness of combining phenotypic, gene expression, and population genetics analyses to uncover key regulatory genes and functional variations (Li et al., 2023).
Additionally, the integration of chromatin accessibility profiles and transcription factor occupancy data has provided insights into the regulatory landscape of maize. For example, profiling accessible chromatin and nucleosome occupancy during early reproductive development revealed regulatory elements that contribute to organogenesis and tissue-specific regulation. This approach has identified new SNP-trait associations in known regulators of inflorescence development, as well as new candidate genes, thereby enhancing our understanding of the cis-regulatory landscape in maize (Parvathaneni et al., 2019).
5.3 Identification of key regulatory networks in maize
The identification of key regulatory networks is crucial for understanding the complex gene regulatory mechanisms in maize. Gene regulatory networks (GRNs) link transcription factors (TFs) to their target genes, representing maps of potential transcriptional regulation. By analyzing a large number of maize transcriptome datasets, researchers have generated multiple coexpression-based GRNs that capture distinct TF-target associations and biological processes. These networks have identified putative regulators of important metabolic pathways and provided potential targets for breeding or biotechnological applications (Zhou et al., 2020).
Furthermore, the integration of chromatin characteristics, such as DNA methylation, chromatin accessibility, and histone modifications, has enabled the prediction of distal enhancer candidates in maize. These enhancer candidates, which display tissue-specificity and regulatory functions, have been validated through chromatin profiling and gene expression analyses. This approach has expanded the toolbox for functional characterization of gene regulation in maize, highlighting the importance of long-range cis-regulatory elements in controlling gene expression and agronomic traits (Oka et al., 2017; Parvathaneni et al., 2020).
In conclusion, advancements in transcriptome sequencing technologies, functional genomics studies, and the identification of key regulatory networks have significantly enhanced researchers understanding of gene expression and regulation in maize. These insights are crucial for improving maize breeding strategies and developing high-yielding, stress-tolerant maize cultivars.
6 Epigenetics and Its Role in Maize Breeding
6.1 Introduction to epigenetic mechanisms in plants
Epigenetics refers to heritable changes in gene expression that do not involve alterations in the DNA sequence itself. These changes are primarily mediated through mechanisms such as DNA methylation, histone modifications, and RNA-directed DNA methylation (RdDM) (Samantara et al., 2021; Gupta and Salgotra, 2022; Tonosaki et al., 2022). In plants, these epigenetic modifications play a crucial role in regulating gene expression, thereby influencing plant development, stress responses, and phenotypic plasticity (Iwasaki and Paszkowski, 2014; Kakoulidou et al., 2021). The concept of epigenetic memory, where chromatin states are propagated through cell divisions, allows plants to retain information about past environmental conditions and adapt accordingly (Iwasaki and Paszkowski, 2014; Huang et al., 2017).
6.2 Epigenetic modifications and their impact on trait expression
Epigenetic modifications can significantly impact the expression of agronomic traits in maize. DNA methylation, one of the primary epigenetic mechanisms, involves the addition of methyl groups to cytosine residues in DNA, leading to changes in chromatin structure and gene expression (Agarwal et al., 2020; Gupta and Salgotra, 2022). Histone modifications, such as acetylation, methylation, and phosphorylation, also play a critical role in regulating chromatin dynamics and gene activity (Duan et al., 2018; Samantara et al., 2021). These modifications can either activate or repress gene expression, depending on the specific chemical groups added to the histones.
In maize, epigenetic regulation has been shown to influence various traits, including stress tolerance, growth, and development. For instance, DNA methylation patterns can change in response to abiotic stresses such as drought, salinity, and heat, leading to the activation of stress-responsive genes (Akhter et al., 2021). These stress-induced epigenetic changes can be stable and heritable, providing a mechanism for plants to adapt to changing environmental conditions over generations (Delcuve et al., 2009; Akhter et al., 2021).
6.3 Potential of epigenetic tools in crop improvement
The potential of epigenetic tools in crop improvement is immense, particularly in the context of global climate change and the need for sustainable agriculture. Epigenetic modifications can be harnessed to develop crops with enhanced traits such as increased yield, stress tolerance, and disease resistance (Kakoulidou et al., 2021; Gupta and Salgotra, 2022). One promising approach is epibreeding, which involves the selection and manipulation of epigenetic variants (epialleles) to achieve desired phenotypes (Gupta and Salgotra, 2022).
Recent advancements in high-throughput sequencing technologies have enabled the comprehensive analysis of plant epigenomes, facilitating the identification of beneficial epigenetic modifications (Huang et al., 2017; Tonosaki et al., 2022). Epigenome editing, which involves targeted modifications of the epigenome using tools such as CRISPR/dCas9, holds great promise for precise and efficient crop improvement (Kakoulidou et al., 2021; Tonosaki et al., 2022). By targeting specific genes or regulatory regions, researchers can modulate gene expression and create new phenotypes without altering the underlying DNA sequence.
Moreover, the integration of epigenetic information into predictive models can enhance the accuracy of breeding programs. By considering both genetic and epigenetic data, breeders can better predict plant performance and select for traits that confer resilience to environmental stresses (Agarwal et al., 2020; Kakoulidou et al., 2021). This holistic approach can accelerate the development of climate-smart crops that are better equipped to thrive in diverse and challenging environments.
In conclusion, the field of epigenetics offers valuable insights and tools for maize breeding and crop improvement. By leveraging epigenetic mechanisms, researchers can unlock new avenues for enhancing crop productivity and sustainability, ultimately contributing to global food security in the face of climate change. The continued exploration and application of epigenetic knowledge will be crucial for the future of agriculture.
7 Future Directions and Prospects
7.1 Emerging genomic technologies and their potential
The field of maize genomics is rapidly evolving, with several emerging technologies poised to revolutionize crop breeding. One of the most promising advancements is the development of genomics-assisted breeding (GAB), which leverages modern genome resources to enhance germplasm and develop new cultivars. GAB 2.0, the next iteration of this technology, aims to fast-track the manipulation of allelic variation, creating novel diversity and facilitating its rapid incorporation into crop improvement programs (Varshney et al., 2021). This approach is expected to play a crucial role in breeding climate-smart maize cultivars with higher nutritional value in a cost-effective and timely manner.
Another significant advancement is genomic selection (GS), which accelerates the breeding cycle by enabling the rapid selection of superior genotypes. GS has shown tangible genetic gains in maize breeding and is expected to further enhance germplasm by integrating genes from gene bank accessions into elite lines (Crossa et al., 2017). The integration of hyperspectral imaging technology with GS and pedigree-assisted breeding could further improve the accuracy and efficiency of these breeding programs.
High-throughput sequencing and re-sequencing technologies are also transforming maize genomics. These technologies allow for the detailed characterization of plant genomes and genetic diversity, facilitating the association of genetic variation with diverse agronomic phenotypes (Bevan et al., 2017). The ability to perform high-throughput resequencing provides opportunities for comparative genomics, which can accelerate crop improvement by identifying and utilizing beneficial genetic variations (Morrell et al., 2011).
CRISPR/Cas9-mediated genome editing is another groundbreaking technology that has heightened the demand for higher transformation efficiencies in maize. This technology allows for precise, site-specific mutagenesis, enabling breeders to obtain desired gene sequences and make sophisticated genomic modifications (Kausch et al., 2021). The integration of CRISPR/Cas9 with advanced genomics and accelerated breeding techniques is expected to significantly contribute to global food security.
7.2 Challenges and limitations in maize genomics
Despite the promising advancements, several challenges and limitations persist in maize genomics. One of the primary challenges is the complexity of the maize genome, which is large and highly repetitive. This complexity makes genome assembly and annotation difficult, hindering the identification and utilization of beneficial genetic variations (Bevan et al., 2017).
Another significant challenge is the integration of genomic data with phenotypic data. While high-throughput phenotyping platforms are available, the accurate and efficient integration of these data sets remains a bottleneck. This integration is crucial for discovering marker-trait associations and optimizing breeding strategies (Barabaschi et al., 2016).
Climate change poses another challenge, as it affects both the quantity and quality of maize crops. Developing new cultivars that are resistant to climate stress without diminishing yield or quality is a pressing need. However, the genetic basis of climate resilience is complex and not fully understood, making it difficult to breed climate-smart maize cultivars (Muntean et al., 2022).
The high cost and technical expertise required for advanced genomic technologies also limit their widespread adoption. Many breeding programs, especially in developing countries, lack the resources and infrastructure to implement these technologies effectively (Andorf et al., 2019). Additionally, regulatory and public acceptance issues surrounding genetically modified organisms (GMOs) and genome-edited crops pose further challenges to the adoption of these technologies (Kausch et al., 2021).
7.3 Strategic roadmap for future research and breeding innovations
To overcome these challenges and fully realize the potential of emerging genomic technologies, a strategic roadmap for future research and breeding innovations is essential. Efforts should be made to simplify and streamline genome assembly and annotation processes. Developing more efficient algorithms and computational tools can help manage the complexity of the maize genome and facilitate the identification of beneficial genetic variations (Bevan et al., 2017).
Integrating genomic data with phenotypic data should be prioritized. Advanced data integration platforms and machine learning algorithms can enhance the accuracy and efficiency of this process, enabling the discovery of marker-trait associations and optimizing breeding strategies (Barabaschi et al., 2016). Collaborative efforts between genomic researchers and breeders can also facilitate the practical application of these integrated data sets in breeding programs.
Addressing climate change requires a multifaceted approach. Research should focus on understanding the genetic basis of climate resilience and identifying key genes and pathways involved in stress tolerance. This knowledge can then be applied to develop new cultivars that are resistant to climate stress without compromising yield or quality (Muntean et al., 2022). Additionally, breeding programs should incorporate climate resilience as a key criterion in their selection processes.
To make advanced genomic technologies more accessible, efforts should be made to reduce costs and build technical capacity in breeding programs, especially in developing countries. This can be achieved through international collaborations, funding support, and training programs that equip breeders with the necessary skills and resources (Andorf et al., 2019). Public engagement and transparent communication about the benefits and safety of GMOs and genome-edited crops can also help address regulatory and acceptance issues (Kausch et al., 2021).
In conclusion, the future of maize genomics holds immense potential for enhancing crop breeding and ensuring global food security. By addressing the current challenges and strategically leveraging emerging technologies, we can pave the way for innovative breeding solutions that meet the demands of a growing population and a changing climate.
8 Concluding Remarks
Recent advancements in maize genomic tools have significantly transformed the landscape of crop breeding. The integration of high-throughput sequencing technologies, such as Single Molecule Real-Time (SMRT) sequencing, has led to the development of more complete and accurate reference genomes, which are crucial for understanding genetic and functional variations in maize. The advent of CRISPR/Cas9 genome editing has enabled precise modifications in maize genomes, facilitating the improvement of complex traits such as yield, drought tolerance, and disease resistance. Additionally, the development of multiplex genome editing strategies, like BREEDIT, has allowed for the simultaneous editing of multiple genes, further accelerating the breeding process. The use of bioinformatics and data sciences has also played a pivotal role in managing the large datasets generated from genomic studies, thereby enhancing the efficiency of breeding programs.
Future research in maize genomics is poised to focus on several key areas. One major direction is the improvement of genome editing specificity and efficiency, particularly through advancements in delivery systems and homology-directed repair mechanisms. Another critical area is the exploration of genetic diversity in wild relatives and landraces of maize to identify novel genetic variations that can be harnessed for crop improvement. The integration of genomic selection with high-throughput phenotyping and rapid generation advancement techniques is expected to further accelerate the breeding cycle and enhance genetic gains. Additionally, there is a growing interest in the application of synthetic biology to create new traits and improve existing ones, which could revolutionize maize breeding.
The advancements in maize genomic tools have profound implications for global agriculture. By enabling the development of high-performance maize cultivars with improved traits, these technologies can significantly contribute to food security, especially in the face of a growing global population and climate change. The ability to rapidly develop drought-tolerant and disease-resistant maize varieties is particularly crucial for sustaining agricultural productivity in tropical and subtropical regions. Moreover, the precision and efficiency of modern breeding techniques can help reduce the time and resources required to develop new cultivars, making it possible to meet the increasing demand for food more sustainably. Overall, the continued advancement and application of maize genomic tools hold great promise for enhancing crop resilience, productivity, and sustainability on a global scale.
Acknowledgments
The authors gratefully acknowledges the valuable feedback provided by two anonymous reviewers, whose insights significantly improved the manuscript.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Agarwal A., Yadava P., Kumar K., Singh I., Kaul T., Pattanayak A., and Agrawal P., 2018, Insights into maize genome editing via CRISPR/Cas9, Physiology and Molecular Biology of Plants, 24: 175-183.
https://doi.org/10.1007/s12298-017-0502-3
PMid:29515313 PMCid:PMC5834987
Agarwal G., Kudapa H., Ramalingam A., Choudhary D., Sinha P., Garg V., Singh V., Patil G., Pandey M., Nguyen H., Guo B., Sunkar R., Niederhuth C., and Varshney R., 2020, Epigenetics and epigenomics: underlying mechanisms, relevance, and implications in crop improvement, Functional and Integrative Genomics, 20: 739-761.
https://doi.org/10.1007/s10142-020-00756-7
PMid:33089419
Akhter Z., Bi Z., Ali K., Sun C., Fiaz S., Haider F., and Bai J., 2021, In response to abiotic stress, DNA methylation confers epigenetic changes in plants, Plants, 10(6): 1096.
Andorf C., Beavis W., Hufford M., Smith S., Suza W., Wang K., Woodhouse M., Yu J., and Lübberstedt T., 2019, Technological advances in maize breeding: past, present and future, Theoretical and Applied Genetics, 132: 817-849.
https://doi.org/10.1007/s00122-019-03306-3
PMid:30798332
Barabaschi D., Tondelli A., Desiderio F., Volante A., Vaccino P., Valè G., and Cattivelli L., 2016, Next generation breeding, Plant Science, 242: 3-13.
https://doi.org/10.1016/j.plantsci.2015.07.010
PMid:26566820
Bevan M., Uauy C., Wulff B., Zhou J., Krasileva K., and Clark M., 2017, Genomic innovation for crop improvement, Nature, 543: 346-354.
https://doi.org/10.1038/nature22011.
PMid:28300107
Bortesi L., and Fischer R., 2015, The CRISPR/Cas9 system for plant genome editing and beyond, Biotechnology Advances, 33(1): 41-52.
https://doi.org/10.1016/j.biotechadv.2014.12.006.
PMid:25536441
Budhlakoti N., Kushwaha A., Rai A., Chaturvedi K., Kumar A., Pradhan A., Kumar U., Kumar R., Juliana P., Mishra D., and Kumar S., 2022, Genomic selection: a tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops, Frontiers in Genetics, 13: 832153.
https://doi.org/10.3389/fgene.2022.832153
PMid:35222548 PMCid:PMC8864149
Crossa J., Pérez-Rodríguez P., Cuevas J., Montesinos-López O., Jarquín D., Campos G., Burgueño J., González-Camacho J., Pérez-Elizalde S., Beyene Y., Dreisigacker S., Singh R., Zhang X., Gowda M., Roorkiwal M., Rutkoski J., and Varshney R., 2017, Genomic selection in plant breeding: methods, models, and perspectives, Trends in Plant Science, 22(11): 961-975.
https://doi.org/10.1016/j.tplants.2017.08.011
PMid:28965742
Delcuve G., Rastegar M., and Davie J., 2009, Epigenetic control, Journal of Cellular Physiology, 219(2): 243-250.
https://doi.org/10.1002/jcp.21678
PMid:19127539
Duan C., Zhu J., and Cao X., 2018, Retrospective and perspective of plant epigenetics in China, Journal of Genetics and Genomics, 45(11): 621-638.
https://doi.org/10.1016/j.jgg.2018.09.004
PMid:30455036
Eathington S., Crosbie T., Edwards M., Reiter R., and Bull J., 2007, Molecular markers in a commercial breeding program, Crop Science, 47: S-154.
https://doi.org/10.2135/cropsci2007.04.0015IPBS
Gupta C., and Salgotra R., 2022, Epigenetics and its role in effecting agronomical traits, Frontiers in Plant Science, 13: 925688..
https://doi.org/10.3389/fpls.2022.925688
PMid:36046583 PMCid:PMC9421166
Hasan N., Choudhary S., Naaz N., Sharma N., and Laskar R., 2021, Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes, Journal of Genetic Engineering and Biotechnology, 19(1): 128..
https://doi.org/10.1186/s43141-021-00231-1
PMid:34448979 PMCid:PMC8397809
He J., Zhao X., Laroche A., Lu Z., Liu H., and Li Z., 2014, Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding, Frontiers in Plant Science, 5: 484.
https://doi.org/10.3389/fpls.2014.00484
PMid:25324846 PMCid:PMC4179701
Huang J., Lynn J., Schulte L., Vendramin S., and McGinnis K., 2017, Epigenetic control of gene expression in maize, International Review of Cell and Molecular Biology, 328: 25-48.
https://doi.org/10.1016/bs.ircmb.2016.08.002
PMid:28069135
Iwasaki M., and Paszkowski J., 2014, Epigenetic memory in plants, The EMBO Journal, 33: 1987-1998.
https://doi.org/10.15252/embj.201488883
PMid:25104823 PMCid:PMC4195768
Jannink J., Lorenz A., and Iwata H., 2010, Genomic selection in plant breeding: from theory to practice, Briefings in Functional Genomics, 9(2): 166-177.
https://doi.org/10.1093/bfgp/elq001
PMid:20156985
Jiao Y., Peluso P., Shi J., Liang T., Stitzer M., Wang B., Campbell M., Stein J., Wei X., Chin C., Guill K., Regulski M., Kumari S., Olson A., Gent J., Schneider K., Wolfgruber T., May M., Springer N., Antoniou E., McCombie W., Presting G., McMullen M., Ross-Ibarra J., Dawe R., Hastie A., Rank D., and Ware D., 2017, Improved maize reference genome with single-molecule technologies, Nature, 546: 524-527.
https://doi.org/10.1038/nature22971
PMid:28605751 PMCid:PMC7052699
Kakoulidou I., Avramidou E., Baránek M., Brunel-Muguet S., Farrona S., Johannes F., Kaiserli E., Lieberman-Lazarovich M., Martinelli F., Mladenov V., Testillano P., Vassileva V., and Maury S., 2021, Epigenetics for crop improvement in times of global change, Biology, 10(8): 766.
Kausch A., Wang K., Kaeppler H., and Gordon-Kamm W., 2021, Maize transformation: history, progress, and perspectives, Molecular Breeding, 41: 1-36.
https://doi.org/10.1007/s11032-021-01225-0
PMid:37309443 PMCid:PMC10236110
Kumlehn J., Pietralla J., Hensel G., Pacher M., and Puchta H., 2018, The CRISPR/Cas revolution continues: From efficient gene editing for crop breeding to plant synthetic biology, Journal of Integrative Plant Biology, 60(12): 1127-1153.
https://doi.org/10.1111/jipb.12734
PMid:30387552
Li C., Brant E., Budak H., and Zhang B., 2021, CRISPR/Cas: a nobel prize award-winning precise genome editing technology for gene therapy and crop improvement, Journal of Zhejiang University Science B, 22: 253-284.
https://doi.org/10.1631/jzus.B2100009
PMid:33835761 PMCid:PMC8042526
Li C., Li Y., Song G., Yang D., Xia Z., Sun C., Zhao Y., Hou M., Zhang M., Qi Z., Wang B., and Wang H., 2023, Gene expression and eQTL analyses uncover natural variations underlying improvement of important agronomic traits during modern maize breeding, The Plant Journal, 115(3): 772-787.
https://doi.org/10.1111/tpj.16260
PMid:37186341
Li C., Song W., Luo Y., Gao S., Zhang R., Shi Z., Wang X., Wang R., Wang F., Wang J., Zhao Y., Su A., Wang S., Li X., Luo M., Wang S., Zhang Y., Ge J., Tan X., Yuan Y., Bi X., He H., Yan J., Wang Y., Hu S., and Zhao J., 2019, The HuangZaoSi maize genome provides insights into genomic variation and improvement history of maize, Molecular Plant, 12(3): 402-409.
https://doi.org/10.1016/j.molp.2019.02.009
PMid:30807824
Manghwar H., Lindsey K., Zhang X., and Jin S., 2019, CRISPR/Cas system: recent advances and future prospects for genome editing, Trends in Plant Science, 24(12): 1102-1125.
https://doi.org/10.1016/j.tplants.2019.09.006
PMid:31727474
Merrick L., Herr A., Sandhu K., Lozada D., and Carter A., 2022, Optimizing plant breeding programs for genomic selection, Agronomy, 12(3): 714.
https://doi.org/10.20944/preprints202202.0048.v1
Monsur M., Shao G., Lv Y., Ahmad S., Wei X., Hu P., and Tang S., 2020, Base editing: the ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants, Genes, 11(4): 466.
https://doi.org/10.3390/genes11040466
Moon S., Kim D., Ko J., and Kim Y., 2019, Recent advances in the CRISPR genome editing tool set, Experimental and Molecular Medicine, 51(11): 1-11.
https://doi.org/10.1038/s12276-019-0339-7
Morrell, P., Buckler, E., and Ross-Ibarra, J., 2011, Crop genomics: advances and applications, Nature Reviews Genetics, 13: 85-96.
https://doi.org/10.1038/nrg3097
Muntean L., ONA A., Berindean I., Racz I., and Muntean S., 2022, Maize breeding: from domestication to genomic tools, Agronomy, 12(10): 2365.
https://doi.org/10.3390/agronomy12102365
Nuss E., and Tanumihardjo S., 2010, Maize: a paramount staple crop in the context of global nutrition, Comprehensive Reviews in Food Science and Food Safety, 9(4): 417-436.
https://doi.org/10.1111/j.1541-4337.2010.00117.x
Oka R., Zicola J., Weber B., Anderson S., Hodgman C., Gent J., Wesselink J., Springer N., Hoefsloot H., Turck F., and Stam M., 2017, Genome-wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize, Genome Biology, 18: 1-24.
https://doi.org/10.1186/s13059-017-1273-4
Ortiz R., Taba S., Tovar V., Mezzalama M., Xu Y., Yan J., and Crouch J., 2010, Conserving and enhancing maize genetic resources as global public goods-a perspective from CIMMYT, Crop Science, 50: 13-28.
https://doi.org/10.2135/cropsci2009.06.0297
Palacios-Rojas N., McCulley L., Kaeppler M., Titcomb T., Gunaratna N., Lopez-Ridaura S., and Tanumihardjo S., 2020, Mining maize diversity and improving its nutritional aspects within agro-food systems, Comprehensive Reviews in Food Science and Food Safety, 19(4): 1809-1834.
https://doi.org/10.1111/1541-4337.12552
PMid:33337075
Parvathaneni R., Bertolini E., Shamimuzzaman M., Vera D., Lung P., Rice B., Zhang J., Brown P., Lipka A., Bass H., and Eveland A., 2019, The regulatory landscape of early maize inflorescence development, Genome Biology, 21: 1-33.
https://doi.org/10.1186/s13059-020-02070-8
PMid:32631399 PMCid:PMC7336428
Ran F., Hsu P., Wright J., Agarwala V., Agarwala V., Scott D., and Zhang F., 2013, Genome engineering using the CRISPR-Cas9 system, Nature Protocols, 8: 2281-2308.
https://doi.org/10.1038/nprot.2013.143
Rice B., and Lipka A., 2021, Diversifying maize genomic selection models, Molecular Breeding, 41(5): 33.
https://doi.org/10.1007/s11032-021-01221-4
PMid:37309328 PMCid:PMC10236107
Romay M., Millard M., Glaubitz J., Peiffer J., Swarts K., Casstevens T., Elshire R., Acharya C., Mitchell S., Flint-Garcia S., McMullen M., Holland J., Buckler E., and Gardner C., 2013, Comprehensive genotyping of the USA national maize inbred seed bank, Genome Biology, 14: 1-18.
https://doi.org/10.1186/gb-2013-14-6-r55
Samantara K., Shiv A., Sousa L., Sandhu K., Priyadarshini P., and Mohapatra S., 2021, A comprehensive review on epigenetic mechanisms and application of epigenetic modifications for crop improvement, Environmental and Experimental Botany, 188: 104479.
https://doi.org/10.1016/j.envexpbot.2021.104479
Thudi M., Palakurthi R., Schnable J., Chitikineni A., Dreisigacker S., Mace E., Srivastava R., Satyavathi C., Odeny D., Tiwari V., Lam H., Hong Y., Singh V., Li G., Xu Y., Chen X., Kaila S., Nguyen H., Sivasankar S., Jackson S., Close T., Shubo W., and Varshney R., 2020, Genomic resources in plant breeding for sustainable agriculture, Journal of Plant Physiology, 257: 153351.
https://doi.org/10.1016/j.jplph.2020.153351
Tonosaki K., Fujimoto R., Dennis E., Raboy V., and Osabe K., 2022, Will epigenetics be a key player in crop breeding? Frontiers in Plant Science, 13: 958359.
https://doi.org/10.3389/fpls.2022.958350
Varshney R., Bohra A., Yu J., Graner A., Zhang Q., and Sorrells M., 2021, Designing future crops: genomics-assisted breeding comes of age, Trends in Plant Science, 26(6): 631-649.
https://doi.org/10.1016/j.tplants.2021.03.010
Varshney R., Mohan S., Gaur P., Gangarao N., Pandey M., Pandey M., Bohra A., Sawargaonkar S., Chitikineni A., Kimurto P., Janila P., Saxena K., Fikre A., Sharma M., Rathore A., Pratap A., Tripathi S., Datta S., Chaturvedi S., Mallikarjuna N., Anuradha G., Babbar A., Choudhary A., Mhase M., Bharadwaj C., Mannur D., Harer P., Guo B., Liang X., Nadarajan N., and Gowda C., 2013, Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics, Biotechnology Advances, 31(8): 1120-1134.
https://doi.org/10.1016/j.biotechadv.2013.01.001
Wan X., Wu S., Li Z., Dong Z., An X., Ma B., Tian Y., and Li J., 2019, Maize genic male-sterility genes and their applications in hybrid breeding: progress and perspectives, Molecular Plant, 12(3): 321-342.
https://doi.org/10.1016/j.molp.2019.01.014
Wang J., and Doudna J., 2023, CRISPR technology: A decade of genome editing is only the beginning, Science, 379(6629): eadd8643.
https://doi.org/10.1126/science.add8643
Zhang M., Kong D., and Wang H., 2023, Genomic landscape of maize domestication and breeding improvement, Seed Biology, 2(1): 9.
https://doi.org/10.48130/seedbio-2023-0009
Zhou P., Hirsch C., Briggs S., and Springer N., 2019, Dynamic patterns of gene expression additivity and regulatory variation throughout maize development, Molecular Plant, 12(3): 410-425.
https://doi.org/10.1016/j.molp.2018.12.015
Zhou P., Li Z., Magnusson E., Cano F., Crisp P., Noshay J., Grotewold E., Hirsch C., Briggs S., and Springer N., 2020, Meta gene regulatory networks in maize highlight functionally relevant regulatory interactions, Plant Cell, 32: 1377-1396.
https://doi.org/10.1105/tpc.20.00080

. PDF(1262KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaojing Yang
. Baixin Song
Related articles
. Maize genomics
. CRISPR/Cas9
. Genomic selection
. High-Throughput phenotyping
. Crop breeding
Tools
. Email to a friend
. Post a comment


