 Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 4 doi: 10.5376/mgg.2024.15.0016
Received: 18 May, 2024 Accepted: 20 Jun., 2024 Published: 05 Jul., 2024
Li J.S., 2024, Chloroplast Genome studies in zea: insights into maize domestication, Maize Genomics and Genetics, 15(4): 160-170 (doi: 10.5376/mgg.2024.15.0016)
This study aims to synthesize current knowledge on the chloroplast genome of Zea genus, with a focus on its significance in understanding maize domestication. By examining the structure, function, and comparative genomics of the chloroplast genome, we elucidate the genetic and evolutionary mechanisms underlying the transition from wild teosinte to cultivated maize. Key findings from chloroplast genome studies reveal that maize originated in the Balsas River Valley, highlighting the genetic diversity within the genus Zea and the evolutionary relationships among maize varieties. Chloroplast DNA analysis uncovers patterns of gene flow, hybridization, and introgression, which contribute to the genetic richness of maize. Positive selection on chloroplast genes associated with photosynthesis and stress response underscores the adaptive changes during domestication. Additionally, advancements in genomic technologies and bioinformatics have enhanced the resolution and accuracy of chloroplast genome assembly and analysis. Chloroplast genome studies provide crucial insights into the genetic and evolutionary dynamics of maize domestication. The integration of chloroplast and nuclear genome data offers a comprehensive understanding of the selective pressures and adaptations that have shaped modern maize. These findings have significant implications for maize breeding, conservation, and global food security, emphasizing the importance of continued research and collaboration in the field of chloroplast genomics.
1 Introduction
Maize (Zea mays) is one of the most significant cereal crops globally, with a rich history of domestication that dates back approximately 9 000 years. Originating from the wild grass teosinte in the region that is now Mexico, maize has undergone extensive genetic and phenotypic changes through selective breeding by indigenous peoples. This process has resulted in the diverse varieties of maize we see today, which are adapted to a wide range of environmental conditions and agricultural practices (Gui et al., 2020). The domestication of maize involved the selection for traits such as increased cob size, kernel number, and reduced seed dispersal mechanisms, which have significantly enhanced its utility as a staple food crop (Liu et al., 2016). Understanding the domestication process of maize not only provides insights into agricultural history but also helps in improving modern breeding techniques for better yield, resilience, and nutritional value.
Chloroplasts are essential organelles in plant cells responsible for photosynthesis, governing essential processes such as photosynthesis, plant metabolism, and adaptation to environmental stresses. Unlike the nuclear genome, the chloroplast genome is maternally inherited and relatively small, making it a valuable tool for phylogenetic and evolutionary studies. The study of chloroplast genomes in maize is crucial for several reasons. Firstly, chloroplasts play a pivotal role in the C4 photosynthetic pathway, which is highly efficient and contributes to the high productivity of maize (Friso et al., 2010; Zhao et al., 2013). Understanding the chloroplast genome can provide insights into the genetic basis of this efficiency. Secondly, chloroplast genomes are relatively small and conserved, making them useful for phylogenetic studies and species identification (Li et al., 2019a; 2020a). Additionally, variations in the chloroplast genome can affect plant development and stress responses, which are critical for crop improvement and adaptation to changing environmental conditions (Udy et al., 2012; Liu et al., 2019).
The primary goal of this study is to provide a comprehensive overview of the current research status of maize chloroplast genomes, emphasizing their importance in understanding the domestication process. The study will summarize the methods used for sequencing and analyzing maize chloroplast genomes, analyze key findings from recent studies on the structure, function, and evolution of maize chloroplast genomes, and discuss how these findings contribute to a broader understanding of maize domestication. Additionally, the study will discuss the impact of chloroplast genome variation on maize physiology, development, and stress responses, identify gaps in current knowledge, and propose directions for future research. By achieving these objectives, this study will contribute to a deeper understanding of the role of chloroplast genomes in maize biology and their potential applications in crop improvement.
2 The Chloroplast Genome of Zea
2.1 Structure and organization
The chloroplast genome of Zea, like many other plant species, exhibits a typical quadripartite structure. This structure includes a large single copy (LSC) region, a small single copy (SSC) region, and a pair of inverted repeat (IRa and IRb) regions. Such organization is consistent with the chloroplast genomes of other species within the Poaceae family and beyond (Li et al., 2019a; 2020a; Yang et al., 2022). The LSC and SSC regions are separated by the IR regions, which are generally more conserved compared to the single copy regions (Biju et al., 2019). This structural organization is crucial for maintaining the stability and functionality of the chloroplast genome.
In maize, the chloroplast genome encodes approximately 110~130 genes, including those essential for photosynthesis (e.g., rbcL, psaA, psbA) and genes involved in transcription and translation (e.g., rpoB, rpoC1). The organization of these genes is highly conserved among angiosperms, with variations in non-coding regions contributing to interspecies diversity.
2.2 Function and significance
The chloroplast genome plays a vital role in photosynthesis and other essential metabolic processes. It encodes genes involved in the photosynthetic machinery, including those for the photosystem I and II complexes, ATP synthase, and the cytochrome b6f complex (Li et al., 2020a; 2020b). These genes are vital for the conversion of light energy into chemical energy, enabling the plant to produce the carbohydrates necessary for growth and development.
Additionally, the chloroplast genome contains genes for ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and various proteins necessary for chloroplast function and maintenance. The presence of these genes underscores the significance of the chloroplast genome in sustaining the energy requirements of the plant through photosynthesis (Li et al., 2019b; Orton et al., 2020).
2.3 Comparison with other plant species
When comparing the chloroplast genome of Zea with those of other plant species, several similarities and differences can be observed. The overall structure, including the presence of LSC, SSC, and IR regions, is highly conserved across different species, such as those in the Zingiberaceae family (Li et al., 2019a; 2020b; Yang et al., 2022). Studies have found that the chloroplast genes of maize, rice, and wheat evolve at similar rates among grass species, with photosynthesis genes undergoing strong purifying selection (Matsuoka et al., 2002). However, variations in the size of these regions and the presence or absence of certain genes can occur. For instance, the absence of the rps19 gene in some Zingiberaceae species due to the expansion of the LSC region highlights the dynamic nature of chloroplast genome evolution (Yang et al., 2022). Additionally, the identification of highly divergent regions and single nucleotide polymorphisms (SNPs) in various species provides valuable markers for phylogenetic studies and species identification (Li et al., 2020a; 2020b; Yang et al., 2022).
The chloroplast genome of Zea is structurally and functionally similar to those of other plant species, with specific variations that contribute to its unique evolutionary path. These insights into the chloroplast genome of Zea enhance understanding of maize domestication and its adaptation to different environments.
3 Techniques for Studying Chloroplast Genomes
3.1 DNA extraction and sequencing
The extraction of high-quality chloroplast DNA (cpDNA) is a critical step in chloroplast genome studies. Traditional methods often involve tissue grinding and homogenization, which can lead to contamination with nuclear DNA and cell debris. A novel protocol has been developed that uses enzyme digestion of tiny leaf strips to separate protoplasts from leaf tissue, thereby protecting chloroplasts from damage and contamination. This method has been successfully applied to various crops, including maize, resulting in high-quality cpDNA suitable for whole-genome sequencing (Liu et al., 2019). High-throughput sequencing technologies, such as those used in the study of Chlamydomonas reinhardtii, have further improved the accuracy and efficiency of chloroplast genome sequencing by providing high coverage and correcting errors in previous genome sequences (Gallaher et al., 2018).
3.2 Bioinformatics and genomic analysis
Bioinformatics tools and genomic analysis are essential for annotating and understanding chloroplast genomes. For instance, a machine learning-based bioinformatics pipeline, DenovoAS_Finder, was developed to annotate transcriptomes without a complete reference genome, achieving an accuracy of up to 91% (Li et al., 2021). This pipeline is particularly useful for complex genomes like those of maize and its wild relatives. Additionally, RNA-Seq data can be used to guide gene annotation, quantify gene expression, and identify post-transcriptional modifications, as demonstrated in the study of Chlamydomonas reinhardtii (Gallaher et al., 2018). Comparative assessments have shown that chloroplast genomes assembled from RNA-Seq data are highly reliable and similar to those obtained from genomic DNA libraries, making RNA-Seq a viable alternative for chloroplast genome assembly (Osuna-Mascaró et al., 2018).
3.3 Comparative genomics
Comparative genomics involves the analysis of chloroplast genomes across different species or within species to understand evolutionary relationships and genetic diversity. In maize, comparative genomics has revealed significant insights into the domestication process. For example, the study of South American maize landraces showed that chloroplast lineages parallel the geographic structuring of nuclear gene pools, indicating distinct evolutionary paths for Andean and lowland South American maize (López et al., 2021). The use of high-throughput sequencing and bioinformatics tools has enabled the identification of extensive genomic and transcriptomic variations between maize and its wild relative teosinte, providing valuable resources for maize breeding and domestication studies (Li et al., 2021).
By integrating advanced DNA extraction methods, high-throughput sequencing technologies, and sophisticated bioinformatics tools, researchers can gain deeper insights into the chloroplast genomes of maize and related species, thereby enhancing understanding of maize domestication and evolution.
4 Insights into Maize Domestication
4.1 Genetic diversity and phylogenetics
The genetic diversity and phylogenetic relationships within maize (Zea mays) have been extensively studied to understand its domestication process. The availability of genomic data has allowed researchers to identify key genetic variations that differentiate domesticated maize from its wild ancestor, teosinte (Stitzer and Ross-Ibarra, 2018; Chen et al., 2021). For instance, studies have shown that maize underwent significant morphological changes during domestication, such as reduced tillering and seed shattering, which are crucial for its adaptation to agricultural environments (Manchanda et al., 2018). Additionally, DNA methylation patterns have been investigated, revealing differentially methylated regions (DMRs) that correlate with recent selection and may have played a role in gene regulation post-domestication (Xu et al., 2020).
4.2 Origin and evolution of maize
The origin and evolution of maize are rooted in its domestication from teosinte in southern Mexico. Genomic studies have provided insights into the evolutionary timeline and the genetic changes that occurred during this process. The domestication of maize involved the selection of traits that were beneficial for human cultivation, such as larger kernels and a more compact plant structure (Xu et al., 2019). These traits were selected over generations, leading to the maize we know today. It was found that the evolutionary history of maize also includes post-domestication adaptation to diverse environments, allowing it to spread far beyond its original habitat (Manchanda et al., 2018; Xu et al., 2020).
4.3 Population structure and gene flow
The population structure of maize has been shaped by both its domestication and subsequent breeding practices. Genetic analyses have revealed distinct population structures within maize, influenced by gene flow between different maize varieties and their wild relatives. For example, Moreno-Letelier et al. (2020) found through genetic analysis of 29 wild and 43 cultivated maize populations that maize domestication was not a single event but the result of multiple gene flow and selection events (Figure 1). Multiple gene flow events between maize and its wild relatives indicate that the domestication process was long and ongoing. These findings help in conserving wild relatives to maintain genetic diversity for the future.
 Figure 1 Population graph depicting relationships among populations, including migrations events, obtained with TreeMix with 30 673 SNPs (Adopted from Moreno-Letelier et al., 2020) Image caption: The figure reveals migration events and genetic drift among different populations; The branch lengths in the figure are proportional to the degree of genetic drift, and migration events are shown through high deviations in the covariance matrix; The results indicate migration events from a highland mexicana population to maize, from maize to another highland mexicana population, from lowland Jalisco to Z. luxurians, and from maize to a parviglumis population; These findings suggest the existence of complex gene flow during the domestication of maize (Adapted from Moreno-Letelier et al., 2020) |
4.4 Selection pressures and adaptation
Selection pressures during maize domestication and subsequent adaptation to new environments have been a major focus of genomic research. The identification of genomic signatures of selection has provided insights into the adaptive traits that were favored during domestication. For instance, the study of DNA methylation has highlighted the role of epigenetic modifications in maize adaptation, with certain DMRs acting as cis-acting elements that modulate gene regulation (Xu et al., 2020). These findings suggest that both genetic and epigenetic factors have played a role in the adaptation of maize to various environmental conditions, contributing to its widespread cultivation and success as a crop (Manchanda et al., 2018; Xu et al., 2020).
The study of the chloroplast genome and other genomic features in maize has provided valuable insights into its domestication, genetic diversity, population structure, and adaptation. These findings not only enhance understanding of maize evolution but also have implications for crop improvement and agricultural practices.
5 Case Studies and Applications
5.1 Tracing lineage and ancestry
Chloroplast genome studies have proven invaluable in tracing the lineage and ancestry of maize (Zea mays). For instance, the analysis of plastome sequences in South American maize landraces has revealed significant haplotype diversity, which aligns with the geographic structuring of nuclear gene pools. López et al. (2021) used next-generation sequencing technology to analyze the complete chloroplast genomes of 30 South American maize landraces and 3 wild maize species, identifying 124 polymorphic loci. These polymorphic loci were mainly concentrated in regions such as psbE-rps18, petN-rpoB, trnL_UAG-ndhF, and rpoC2-atpI. The study results showed significant differences in haplotype distribution between Andean and South American lowland maize landraces, reflecting the gene pool structure inferred from nuclear markers (Figure 2) (López et al., 2021). These insights are crucial for understanding the domestication and evolutionary history of maize, providing a framework for understanding evolutionary processes at low taxonomic levels and becoming increasingly important for future plant barcoding efforts.
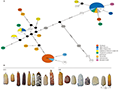 Figure 2 (A) Median-joining network of complete Z. mays plastomes. Circle size is proportional to haplotype frequencies Colours denote category (inbred line, teosinte, ancient sample) or previous group assignment of landraces based on nuclear markers according to Bracco et al. (2016) (Andean, NEA Flours, NEA Popcorns, Tropical Lowland, Highland Mexico and USA). Markers on the lines joining haplotypes represent one mutational step and black circles indicate missing vectors. Edge lengths are not to scale; (B) Morphological diversity in South American landraces carrying H1 and H2 haplotypes (Adopted from López et al., 2021) Image caption: The figure shows that haplotype H1 is most common in NEA Flours maize and also appears in five other groups, but is absent in Andean landraces. Haplotype H2 is predominant in Andean samples and absent in lowland groups. These network relationships reveal the distribution and interrelationships of haplotypes in maize and wild maize species across different groups (Adapted from López et al., 2021) |
5.2 Evolutionary studies
Chloroplast genomes offer a window into the evolutionary processes of plants. In maize, the study of plastome diversity has shown that domesticated maize has distinct chloroplast genomes compared to its wild relatives. This divergence suggests that the domestication process captured significant chloroplast genome diversity, which may have implications for adaptive evolution (Moner et al., 2020). Additionally, comparative analyses of chloroplast genomes in other plant families, such as Ranunculaceae and Zingiberaceae, have provided robust phylogenetic frameworks and clarified long-standing taxonomic controversies (Zhai et al., 2019; Li et al., 2020a).
5.3 Conservation genetics
The conservation of genetic diversity is essential for the resilience and adaptability of plant species. Chloroplast genome studies in maize have highlighted the importance of preserving intraspecific variation. The identification of polymorphic loci within the chloroplast genome can serve as molecular markers for conservation efforts. For example, the study of chloroplast genomes in the Dracunculus clade has identified suitable polymorphic loci that could be used for phylogenetic inference and conservation genetics (Henriquez et al., 2020). Similarly, the comparative analysis of chloroplast genomes in Cleomaceae species has revealed hotspot genes that could be used for species authentication and conservation (Alzahrani et al., 2021).
5.4 Breeding and crop improvement
Chloroplast genome studies have significant applications in breeding and crop improvement. The identification of chloroplast haplotypes and their association with specific traits can inform breeding programs aimed at enhancing crop performance. For instance, the study of chloroplast genomes in rice has shown that different evolutionary paths of cytoplasmic and nuclear genomes have resulted in functional chloroplast genome diversity, which may impact crop performance (Moner et al., 2020). In maize, understanding the distribution of chloroplast haplotypes can aid in the selection of landraces with desirable traits for breeding programs (López et al., 2021). Additionally, the identification of highly divergent regions in chloroplast genomes can provide molecular markers for species identification and phylogenetic studies, facilitating the development of improved crop varieties (Li et al., 2020b).
By leveraging the insights gained from chloroplast genome studies can enhance understanding of maize domestication, evolution, and conservation, ultimately contributing to the development of more resilient and productive crop varieties.
6 Challenges and Limitations
6.1 Technical challenges
One of the primary technical challenges in studying the chloroplast genome of maize (Zea mays) is the complexity of isolating high-purity chloroplast DNA (cpDNA). Traditional methods often involve tissue grinding and homogenization, which can damage chloroplasts and lead to contamination with nuclear DNA and cell debris. This issue has been addressed in other crops like foxtail millet through the development of new protocols that use enzyme digestion to separate protoplasts from leaf tissue, thereby protecting chloroplasts from damage and contamination (Liu et al., 2019). Adapting such protocols for maize could significantly improve the quality of cpDNA and the accuracy of subsequent genomic analyses.
6.2 Data interpretation and complexity
Interpreting the data from chloroplast genome studies in maize presents its own set of challenges. The chloroplast genome of maize, like other plants, contains a large single-copy region (LSC), a small single-copy region (SSC), and two inverted repeat (IR) regions, which complicates the assembly and annotation processes (Chen et al., 2020). Additionally, the presence of distinct clades within the chloroplast genomes of domesticated plants, as seen in rice, suggests that maize may also exhibit significant intra-species diversity that needs to be carefully analyzed (Moner et al., 2020). This diversity can lead to complex phylogenetic relationships that are difficult to resolve, especially when the evolutionary paths of the cytoplasmic and nuclear genomes differ, as observed in rice (Moner et al., 2020).
6.3 Future research directions
Future research in the field of maize chloroplast genomics should focus on several key areas. Improving DNA extraction protocols to ensure high-purity cpDNA will be crucial for obtaining accurate genomic sequences (Liu et al., 2019). Comprehensive phylogenetic studies should be conducted to understand the evolutionary relationships within maize and between maize and its wild relatives. This could involve the use of advanced sequencing technologies and bioinformatics tools to analyze large datasets and resolve complex phylogenetic trees (Chen et al., 2020). Investigating the functional implications of chloroplast genome diversity in maize could provide insights into how different chloroplast types contribute to crop performance and adaptation (Aliyeva et al., 2020; Yang and Yan, 2021). This could involve studying the expression and function of specific genes and proteins that differ between chloroplast clades, as has been suggested for rice (Moner et al., 2020).
By addressing these challenges and focusing on these research directions, researchers can gain a deeper understanding of maize domestication and improve maize breeding programs to enhance crop performance and resilience.
7 Future Perspectives
7.1 Integrating chloroplast and nuclear genomes
The integration of chloroplast and nuclear genome data offers a comprehensive understanding of maize domestication and its evolutionary history. Studies have shown that the evolutionary paths of cytoplasmic and nuclear genomes can differ significantly, as observed in rice, where distinct chloroplast clades were identified (Moner et al., 2020). In maize, leveraging both chloroplast and nuclear genome data can elucidate the complex interactions and evolutionary processes that have shaped modern maize varieties. This integrated approach can also help identify key genetic variations that contribute to important agronomic traits (Li et al., 2021; Zhang et al., 2023).
7.2 Advances in genomic technologies
Recent advancements in genomic technologies, such as single-molecule long-read sequencing and machine learning-based bioinformatics pipelines, have significantly enhanced our ability to study complex genomes like that of maize (Li et al., 2021). These technologies allow for the accurate annotation of transcriptomes and the construction of high-quality genome assemblies, providing deeper insights into the genetic basis of maize domestication and improvement. Additionally, the development of comprehensive databases like ZEAMAP facilitates the integration and visualization of multi-omics data, further accelerating genetic research and breeding efforts (Gui et al., 2020).
7.3 Implications for plant breeding and conservation
The insights gained from chloroplast genome studies have profound implications for plant breeding and conservation. Understanding the genetic diversity within chloroplast genomes can inform breeding strategies aimed at enhancing crop resilience and productivity. For instance, chloroplast metabolic engineering has been proposed as a method to biofortify crops, addressing nutritional deficiencies and improving food security (Tanwar et al., 2022). Moreover, the identification of key genes and regulatory networks involved in stress responses and organ-specific functions can guide the development of maize varieties better adapted to changing environmental conditions (Hoopes et al., 2019).
7.4 Global collaboration and data sharing
Global collaboration and data sharing are essential for advancing our understanding of maize genomics and its application in breeding programs. The establishment of platforms like ZEAMAP, which integrates diverse genomic and phenotypic data, exemplifies the benefits of collaborative efforts in the scientific community (Gui et al., 2020). By sharing data and resources, researchers can build on each other's work, accelerating discoveries and innovations. Furthermore, international partnerships can facilitate the conservation of genetic diversity in maize and its wild relatives, ensuring the sustainability of this vital crop for future generations (Liu et al., 2019).
8 Concluding Remarks
The study of chloroplast genomes in Zea species, particularly maize (Zea mays), has provided significant insights into the domestication and evolutionary history of this crucial crop. Chloroplast genomes have been shown to harbor distinct clades that reflect the domestication events and subsequent diversification of maize. For instance, the chloroplast genomes of domesticated rice, which show distinct clades from their wild relatives, suggest a similar pattern might be observed in maize, indicating multiple domestication events or significant gene flow from wild relatives .
In maize, the chloroplast genome has been sequenced and analyzed, revealing its structure and gene content, which includes 85 protein-coding genes, 25 tRNA genes, and 8 rRNA genes. This comprehensive genomic information is crucial for understanding the functional aspects of chloroplasts in maize, such as their role in photosynthesis and starch metabolism, which are vital for plant development and productivity .
Furthermore, the integration of multi-omics data, including chloroplast genomes, has been facilitated by databases like ZEAMAP, which provide a platform for comparative genomics and the study of domestication signals between maize and its wild relatives. This integration is essential for identifying genetic variations and understanding the evolutionary forces that have shaped the maize genome.
Chloroplast genomes play a pivotal role in elucidating the domestication process of maize. The distinct clades observed in chloroplast genomes can indicate multiple domestication events or significant gene flow from wild relatives, as seen in other crops like rice. In maize, the chloroplast genome has been used to trace the evolutionary history and domestication pathways, revealing the contributions of different teosinte subspecies to the modern maize gene pool .
The chloroplast genome's structure and function are also critical for understanding the physiological adaptations that occurred during domestication. For example, the role of chloroplasts in starch metabolism and photosynthesis is crucial for maize's development and productivity, highlighting the importance of chloroplast-associated genes in the domestication process.
Moreover, the integration of chloroplast genomic data with other omics data through platforms like ZEAMAP allows for a comprehensive understanding of the genetic and phenotypic changes that occurred during domestication. This holistic approach enables researchers to identify key genetic variations and their functional implications, providing insights into how domestication has shaped the maize genome and its agronomic traits.
In conclusion, chloroplast genome studies in Zea species offer valuable insights into the domestication and evolutionary history of maize. By understanding the genetic and functional aspects of chloroplasts, researchers can better comprehend the complex processes that have led to the development of modern maize varieties, ultimately aiding in the improvement of this vital crop.
Acknowledgments
The author extends sincere thanks to two anonymous peer reviewers for their feedback on the manuscript.
Conflict of Interest Disclosure
The author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Aliyeva N., Aliyeva D., Suleymanov S., Rzayev F., Gasimov E., and Huseynova I., 2020, Biochemical properties and ultrastructure of mesophyll and bundle sheath thylakoids from maize (Zea mays) chloroplasts, Functional Plant Biology, 47(11): 970-976.
https://doi.org/10.1071/FP20004
PMid:32574552
Alzahrani D., Albokhari E., Yaradua S., and Abba A., 2021, Complete chloroplast genome sequences of Dipterygium glaucum and Cleome chrysantha and other cleomaceae species, comparative analysis and phylogenetic relationships, Saudi Journal of Biological Sciences, 28: 2476-2490.
Biju V.C., Vijayan S.P.R.S., Vijayan S., Rajan V., Sasi A., Janardhanan A., and Nair A., 2019, The complete chloroplast genome of Trichopus zeylanicus, and phylogenetic analysis with dioscoreales, The Plant Genome, 12(3): 190032.
https://doi.org/10.3835/plantgenome2019.04.0032
PMid:33016590
Bracco M., Cascales J., Hernández J.C., Poggio L., Gottlieb A.M., and Lia V.V., 2016, Dissecting maize diversity in lowland South America: genetic structure and geographic distribution models, BMC Plant Biology, 16: 1-13.
https://doi.org/10.1186/s12870-016-0874-5
PMid:27561710 PMCid:PMC5000442
Chen Y., Ma Y., and Xu X., 2020, The complete chloroplast genome sequence of corn and economic analysis on production costs profits in Zhengzhou city, Mitochondrial DNA Part B Resources, 5(3): 2447-2448.
https://doi.org/10.1080/23802359.2020.1775524
PMid:33457821 PMCid:PMC7782024
Chen Z., Sun J.L., Li D.D., Li P.C., He K., Ali F., Pan Q., Mi G., Chen F., and Yuan L., 2021, Plasticity of root anatomy during domestication of a maize-teosinte derived population, Journal of Experimental Botany, 73(1): 139-153.
https://doi.org/10.1093/jxb/erab406
PMid:34487165
Friso G., Majeran W., Huang M., Sun Q., and Wijk K., 2010, Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly, Plant Physiology, 152(3): 1219-1250.
https://doi.org/10.1104/pp.109.152694
PMid:20089766 PMCid:PMC2832236
Gallaher S., Fitz-Gibbon S., Strenkert D., Purvine S., Pellegrini M., and Merchant S., 2018, High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates, The Plant Journal, 93(3): 545-565.
https://doi.org/10.1111/tpj.13788
PMid:29172250 PMCid:PMC5775909
Gui S., Yang L., Li J., Luo J., Xu X., Yuan J., Chen L., Li W., Yang X., Wu S., Li S., Wang Y., Zhu Y., Gao Q., Yang N., and Yan J., 2020, ZEAMAP, a comprehensive database adapted to the maize multi-omics era, iScience, 23(6): 101241.
https://doi.org/10.1016/j.isci.2020.101241
PMid:32629608 PMCid:PMC7306594
Henriquez C.L., Mehmood F., Hayat A., Sammad A., Waseem S., Waheed M., Matthews P., Croat T., Poczai P., and Ahmed I., 2020, Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae), Genomics, 113(1): 183-192.
https://doi.org/10.1016/j.ygeno.2020.12.016
PMid:33326831
Hoopes G., Hamilton J., Hamilton J., Wood J., Wood J., Esteban E., Pasha A., Vaillancourt B., Vaillancourt B., Provart N., and Buell C., 2019, An updated gene atlas for maize reveals organ‐specific and stress‐induced genes, The Plant Journal, 97: 1154-1167.
https://doi.org/10.1111/tpj.14184
PMid:30537259 PMCid:PMC6850026
Li D., Zhao C., and Liu X., 2019a, Complete Chloroplast genome sequences of kaempferia galanga and kaempferia elegans: molecular structures and comparative analysis, Molecules, 24(3): 474.
https://doi.org/10.3390/molecules24030474
PMid:30699955 PMCid:PMC6385120
Li D.M., Zhu G., Xu Y.C., Ye Y., and Liu J., 2019b, Characterization and phylogenetic analysis of the complete chloroplast genome of Curcuma zedoaria (Zingiberaceae), Mitochondrial DNA Part B, 5: 1329-1331.
https://doi.org/10.1080/23802359.2020.1734496
Li D.M., Ye Y.J., Xu Y.C., Liu J., and Zhu G., 2020a, Complete chloroplast genomes of Zingiber montanum and Zingiber zerumbet: genome structure, comparative and phylogenetic analyses, PLoS One, 15(7): e0236590.
https://doi.org/10.1371/journal.pone.0236590
PMid:32735595 PMCid:PMC7394419
Li D., Zhu G., Xu, Y., Ye Y., and Liu J., 2020b, Complete chloroplast genomes of three medicinal alpinia species: genome organization, comparative analyses and phylogenetic relationships in family zingiberaceae, Plants, 9(2): 286.
https://doi.org/10.3390/plants9020286
PMid:32102387 PMCid:PMC7076362
Li Z., Han L., Luo Z., and Li L., 2021, Single‐molecule long‐read sequencing reveals extensive genomic and transcriptomic variation between maize and its wild relative teosinte (Zea mays ssp. parviglumis), Molecular Ecology Resources, 22: 272-282.
https://doi.org/10.1111/1755-0998.13454
PMid:34157795
Liu D., Cui Y.J., Li S., Bai G., Li Q., Zhao Z., Liang D., Wang C., Wang J., Shi X., Chen C., Feng G., and Liu Z., 2019, A new chloroplast DNA extraction protocol significantly improves the chloroplast genome sequence quality of foxtail millet (Setaria italica (L.) P. Beauv.), Scientific Reports, 9(1): 16227.
https://doi.org/10.1038/s41598-019-52786-2
PMid:31700055 PMCid:PMC6838068
Liu F., Xu Y., Han G., Zhou L., Ali A., Zhu S., and Li X., 2016, Molecular evolution and genetic variation of G2-like transcription factor genes in maize, PLoS One, 11(8): e0161763.
https://doi.org/10.1371/journal.pone.0161763
PMid:27560803 PMCid:PMC4999087
Liu J., Fernie A., and Yan J., 2019, The past, present, and future of maize improvement: domestication, genomics, and functional genomic routes toward crop enhancement, Plant Communications, 1(1): 100010.
https://doi.org/10.1016/j.xplc.2019.100010
PMid:33404535 PMCid:PMC7747985
López M., Fass M., Rivas J., Carbonell-Caballero J., Vera P., Puebla A., Defacio R., Dopazo J., Paniego N., Hopp H., and Lia V., 2021, Plastome genomics in South American maize landraces: chloroplast lineages parallel the geographic structuring of nuclear gene pools, Annals of Botany. 128(1): 115-125.
https://doi.org/10.1093/aob/mcab038
PMid:33693521 PMCid:PMC8318110
Manchanda N., Snodgrass S., Ross-Ibarra J., and Hufford M., 2018, Evolution and adaptation in the maize genome, The Maize Genome, (2018): 319-332.
https://doi.org/10.1007/978-3-319-97427-9_19
Matsuoka Y., Yamazaki Y., Ogihara Y., and Tsunewaki K., 2002, Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals, Molecular Biology and Evolution, 19(12): 2084-2091.
https://doi.org/10.1093/oxfordjournals.molbev.a004033
PMid:12446800
Moner A., Furtado A., and Henry R., 2020, Two divergent chloroplast genome sequence clades captured in the domesticated rice gene pool may have significance for rice production, BMC Plant Biology, 20: 1-9.
https://doi.org/10.1186/s12870-020-02689-6
PMid:33054735 PMCid:PMC7558744
Moreno-Letelier A., Aguirre-Liguori J., Piñero D., Vázquez-Lobo A., and Eguiarte L., 2020, The relevance of gene flow with wild relatives in understanding the domestication process, Royal Society Open Science, 7(4): 191545.
https://doi.org/10.1098/rsos.191545
PMid:32431864 PMCid:PMC7211868
Orton L., Fitzek E., Feng X., Grayburn W., Mower J., Liu K., Zhang C., Duvall M., and Yin Y., 2020, Zygnema circumcarinatum UTEX 1559 chloroplast and mitochondrial genomes provide insight into land plant evolution, Journal of Experimental Botany, 71(11): 3361-3373.
https://doi.org/10.1093/jxb/eraa149
PMid:32206790
Osuna-Mascaró C., Casas R., and Perfectti F., 2018, Comparative assessment shows the reliability of chloroplast genome assembly using RNA-seq, Scientific Reports, 8(1): 17404.
https://doi.org/10.1038/s41598-018-35654-3
PMid:30479362 PMCid:PMC6258696
Stitzer M., and Ross-Ibarra J., 2018, Maize domestication and gene interaction, The New Phytologist, 220(2): 395-408.
https://doi.org/10.1111/nph.15350
PMid:30035321
Tanwar N., Arya S., Rookes J., Cahill D., Lenka S., and Bansal K., 2022, Prospects of chloroplast metabolic engineering for developing nutrient-dense food crops, Critical Reviews in Biotechnology, 43: 1001-1018.
https://doi.org/10.1080/07388551.2022.2092717
PMid:35815847
Udy D., Belcher S., Williams-Carrier R., Gualberto J., and Barkan A., 2012, Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize, Plant Physiology, 160: 1420-1431.
https://doi.org/10.1104/pp.112.204198
PMid:22977281 PMCid:PMC3490597
Wang W., and Lanfear R., 2019, Long-reads reveal that the chloroplast genome exists in two distinct versions in most plants, Genome Biology and Evolution, 11: 3372-3381.
https://doi.org/10.1093/gbe/evz256
PMid:31750905 PMCid:PMC7145664
Xu G., Cao J., Wang X., Chen Q., Jin W., Li Z., and Tian F., 2019, Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor, teosinte, Plant Cell, 31(9): 1990-2009.
https://doi.org/10.1105/tpc.19.00111
PMid:31227559 PMCid:PMC6751114
Xu G., Lyu J., Li Q., Liu H., Wang D., Zhang M., Springer N., Ross-Ibarra J., and Yang J., 2020, Evolutionary and functional genomics of DNA methylation in maize domestication and improvement, Nature Communications, 11(1): 5539.
https://doi.org/10.1038/s41467-020-19333-4
PMid:33139747 PMCid:PMC7606521
Yang N., and Yan J., 2021, New genomic approaches for enhancing maize genetic improvement, Current opinion in Plant Biology, 60: 101977.
https://doi.org/10.1016/j.pbi.2020.11.002
PMid:33418269
Yang Q., Fu G., Wu Z., Li L., Zhao J., and Li Q., 2022, Chloroplast genome evolution in four montane zingiberaceae taxa in China, Frontiers in Plant Science, 12: 774482.
https://doi.org/10.3389/fpls.2021.774482
PMid:35082807 PMCid:PMC8784687
Zhai W., Duan X., Zhang R., Guo C., Li L., Xu G., Shan H., Kong H., and Ren Y., 2019, Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the ranunculaceae, Molecular Phylogenetics and Evolution, 135: 12-21.
https://doi.org/10.1016/j.ympev.2019.02.024
PMid:30826488
Zhang M., Kong D., and Wang H., 2023, Genomic landscape of maize domestication and breeding improvement, Seed Biology, 2: 9.
https://doi.org/10.48130/SeedBio-2023-0009
Zhao Q., Chen S., and Dai S., 2013, C4 photosynthetic machinery: insights from maize chloroplast proteomics, Frontiers in Plant Science, 4: 85.
https://doi.org/10.3389/fpls.2013.00085
Zhu T., Li Z., An X., Long Y., Xue X., Xie K., Ma B., Zhang D., Guan Y., Niu C., Dong Z., Hou Q., Zhao L., Wu S., Li J., Jin W., and Wan X., 2020, Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize, Molecular Plant, 13(11): 1624-1643.
https://doi.org/10.1016/j.molp.2020.09.013
PMid:32956899

. PDF(639KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jiansheng Li
Related articles
. Zea genus
. Chloroplast genome
. Maize domestication
. Genetic diversity
. Phylogenetics
. Gene flow
. Genomic technologies
. Plant breeding
Tools
. Email to a friend
. Post a comment


