This article was first published in Genomics and Applied Biology in Chinese, and here was authorized to translate and publish the paper in English under the terms of Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Rice is one of the main food crops in the world and as the spatle food of 60%~70% in China (Fernandez and Orth, 2018). Rice blast is one of the most serious fungal diseases in rice, A large-scale outbreak of rice blast will cause a significant reduction or even a loss of rice production, seriously affecting the safety of rice production (Martin et al., 2016; Deng et al., 2017). It is estimated that rice destroyed by rice blast is enough to feed 60 million people every year, in the past 20 years, rice blast disease was also present up upward trend (Roychowdhury et al., 2013). It was found that the spores of Magnaporthe grisea were used as the primary and reinfected inoculums, air flow is used as a medium to invade the plant directly from the leaf surface. The initial stages of plant infection by Magnaporthe grisea includes spore adhesion, spore germination, germ tube growth and the formation of infected structures (adherent cells), the pore germination and attachment cell formation play key role in the infection of rice by Magnaporthe grisea (Hamer and Talbot, 1999), The attached bubble can generate huge swelling pressure and will penetrate the nail to break through the cuticle of the rice leaf and quickly infect the rice (Ye et al., 2015; Ali and Mvnir, 2017). In general, keratinase can make plants sick by destroying the cuticle of plant epidermis and catalyzing the hydrolysis of host cell keratinocyte polymers (Duan et al., 2017), However, the same function of keratinase was not found in rice infected by Magnaporthe grisea, We speculated that keratinase may have other biological functions independent of its hydrolase activity in the infection of Magnaporthe grisea.
The results of bioinformatics of MGG_14095 protein showed that the molecular weight of MGG_14095 is 28001.59, the PI was 5.72 and the instability index was 50.99, belongs to acid unstable protein; It contains the conserved domain of keratinase protein belongs to the α/β hydrolase family; It has obvious transmembrane structure and signal peptide cutting site, which belongs to secretory protein. Due to the existence of transm- embrane structure and signal peptide cleavage sites, protein solubility will be reduced, which has a great impact on the purification process, In this experiment, the PCR method was used to clone the MGG_14095 gene except the transmembrane structure and the signal peptide cleavage site, and renamed the MGG_14095 (38-281) gene,selected the prokaryotic expression vector pETM20 with TrxA (thioredoxin protein tag) and His (polyhistidine tag), Construction of pETM20-MGG_14095 (38-281) recombinant system vector, transformed into Escherichia coli DH5α to replicate a large number of recombinant genes, and then transformed into Rosetta (DE3) to express the fusion protein TrxA-MGG_14095 (38-281). After purification by step chromatography, the highly purified soluble target protein was successfully obtained. The result is the analysis of MGG_14095 Protein structure, analysis of the pathogenic mechanism of Magnaporthe grisea provide experimental basis, and provide a theoretical basis for the spread and control of Magnaporthe grisea.
1 Results and Analysis
1.1 Prediction of primary structure and physical and chemical properties of MGG_14095 protein
The MGG_14095 protein is composed of 281 amino acids, of which cysteine accounts for 1.1% of the total number of amino acids (
Figure 1), the molecular mass is 28 001.59, the isoelectric point (pI) is 5.72, and the instability index is 50.99, MGG_14095 protein is belongs to acid unstable protein.
.png) .png)
Figure 1 Primary structure of MGG_14095 protein
Note: A: Alanine; R: Arginine; N: APsaragine; D: APsartic acid; C: Cystine; Q: Glutarnine; E: Glutamicacid; G:Glycine; H: Histidine; I: Isoleucine; L: Leucine; K: Lysine; M: Methionine; F: Phenylalanine; P: Proline; S: Serine;T: Threonine; Y:Tyrosine; V: Valine
|
1.2 MGG_14095 conserved domain prediction of MGG_14095 protein
Use Blast program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to perform homology analysis of cutinase MGG-09100 protein, MGG_14095 protein contains TFIIA transcription factor bound to the promoter, which belongs to α / The β hydrolase superfamily (
Figure 2). Analysis of the conserved domain of the MGG_14095 protein with the pfam program revealed that the protein contains a conserved domain of cutinase.
.png) .png)
Figure 2 Conserved domain prediction of MGG_14095 protein
|
1.3 Prediction of cross-membrane structure and signal peptide position of MGG_14095 protein
The prediction results of the transmembrane structure indicate (
Figure 3) that there may be a significant transmembrane structure from the 7th amino acid to the 35th amino acid of the MGG_14095 protein. Normally, the highest peak of Y value indicates the signal peptide cleavage site (
Figure 4). There is a signal peptide cleavage site between amino acids 25 and 35 of MGG_14095. The MGG_14095 protein is a secreted protein.
.png) .png)
Figure 3 Prediction of cross-membrane structure of MGG_14095 protein
|
.png) 
Figure 4 Prediction of signal peptide position of MGG_14095 protein
|
1.4 MGG_14095(38-281) gene cloning and construction of expression vector
Designed primers to amplify the
MGG_14095 (
38-281) gene (
Figure 5) and amplified by agarose gel electrophoresis. A single band appeared between 500bp and 750bp, which was consistent with the expected
MGG_14095 (
38-281) gene (738 bp) size.
.png) .png)
Figure 5 The amplification result of MGG_14095 (38-281) gene
Note: M: DL 2000 Marker; 1: MGG_14095 (38-281) gene amplification result (with enzyme cut points XhoⅠ); 2: MGG_14095 (38-281) gene amplification result (with enzyme cut points NcoⅠ)
|
The recombinant plasmid was detected by PCR with T7 and t7ter (3 replicates). The positive bands were found at 1 310 (
Figure 6). According to the sequencing results,
mgg_ (
38-281) gene was correctly connected with petm20 vector, and petm20-mgg_ (38-281) vector was successfully constructed.
.png) 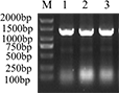
Figure 6 Recombinant plasmid pETM20-MGG_14095(38-281)PCR verification results
Note: M: DL2000 Marker; 1-3: Recombinant plasmid pETM20-MGG_14095(38-281)PCR verification results
|
1.5 Induced expression result and solubility analysis of fusion protein TrxA-MGG_14095(38-281) protein
At 18℃ 0.1 mmol/L IPTG, the fusion protein TrxA-MGG_14095 (38-281) (
Figure 7) was induced and expr- essed. 1 swimlane was the total protein in the uninduced cell at 37℃, and 2th~3th was 0.1 mmol/L After IPTG induction, the expression of TrxA-MGG_14095 (38-281) was obvious. 4th swimlane was protein supernatant after ultrasonic breaking and centrifugation, 5th swimlane was precipitated heavy suspension after ultrasonic breaking and centrifugation. A small part of TrxA-MGG_14095 (38-281) existed in the supernatant in soluble form, most of them in inclusion body form, and most of them in precipitation The fusion protein was purified from the supernatant.
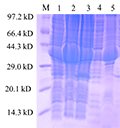
Figure 7 Fusion protein TrxA-MGG_14095(38-281) protein induced expression result
Note: M: Low protein Molecular Weight Marker; 1: The total protein was not induced; 2~3: Total cell protein after induction; 4: Centrifugal crushing after up-clearing; 5: Centrifugal crushing after precipitation
|
1.6 Fusion protein TrxA-MGG_14095 (38-281) affinity purification results
The fusion protein TrxA-MGG_14095(38-281) was eluted with different concentrations of imidazole buffer (
Figure 8). When the concentration of imidazole was 60 mmol/L, the target protein began to be eluted. With the increase of the concentration of imidazole in the buffer, the purity of the target protein gradually increased. When the concentration of imidazole in the buffer was 500 The protein was completely eluted at mmol/L, and the eluates of 100 mmo
l/L, 200 mmo
l/L, 300 mmol/L and 400 mmol/L were collected for ion exchange chromatography.
.png) 
Figure 8 Fusion protein TrxA-MGG_14095 (38-281) affinity purification results
Note: M: low protein Marker; 1: Through the fluid; 2: 40 mmol/L imidazole elution; 3: 60 mmol/L imidazole elution; 4: 80 mmol/L imidazole elution; 5: 100 mmol/L imidazole elution; 6: 200 mmol/L imidazole elution; 7: 300 mmol/L imidazole elution; 8: 400 mmol/L imidazole elution; 9: 500 mmol/L imidazole elution
|
1.7 Fusion protein TrxA-MGG_14095 (38-281) Ion exchange chromatography
The results showed that when the volume of eluate was 16ml, the target protein began to elute, and the charged property of TrxA-MGG_14095 (38-281) was uniform and tolerant to low salt condition (
Figure 9).
.png) 
Figure 9 Fusion protein TrxA-MGG_14095 (38-281) Ion exchange chromatography and SDS-PAGE electrophoresis detection
|
1.8 Fusion protein TrxA-MGG_14095 (38-281) Gel filtration chromatography
After the fusion protein TrxA-MGG_14095 (38-281) protein was subjected to gel filtration chromatography, two protein absorption peaks appeared (
Figure 10). At 9 mL and 14 mL, the corresponding positions were sampled for SDS-PAGE gel verification It was found that all of them had protein bands at the same position, presuming that the protein exists as a dimer.
.png) 
Figure 10 Fusion protein TrxA-MGG_14095 (38-281) Gel filtration chromatography
|
2 Discussion
Cutin is the main component of the cuticle of a plant, and enzymatic hydrolysis of cutin is the first step of rice blast fungus invasion into rice (
Zhang and Xu, 2017). Cutinase is an enzyme secreted by plants when they are stimulated by pathogenic bacteria. It can hydrolyze cutin into monomers to induce more cutinase synthesis and accelerate plant disease (
Qiu and Wang, 2004;
Zhang et al., 2013), When rice blast infects rice, cutinase exists as other biological functions independent of its hydrolase activity, and its specific functions should be further explored. The rice blast fungus MGG_14095 protein contains a conserved domain of cutinase protein, which provides convenient conditions for understanding the role of cutinase in the process of rice blast infection in rice. Before the experiment, bioinformatics analysis was performed on the MGG_14095 protein, we found: 1. MGG_14095 protein contains cysteine, and a small amount of DTT should be added to prevent the formation of intermolecular disulfide bonds during the purification process; 2. Use its charged properties to select an appropriate ion exchange column to help the purification of the target protein. Cation exchange adsorption column combined to help target protein elution 3. MGG_14095 protein is an unstable protein, and it should always be kept at a low temperature during purification.;4. MGG_14095 protein has obvious transmembrane structure and signal peptide cleavage site, which will increase the difficulty of protein purification. The rest of the protein membrane structure and signal peptide cleavage site should be cloned,Based on the above factors, we cloned the rice blast fungus
MGG_14095 (
38-281) gene and selected the pETM20 vector for prokaryotic expression,The pETM20 vector contains TrxA tags and His tags. TrxA tags have a strong translation initiation signal and can drive high-level protein expression (
Chen, 2015). His tags are small enough to minimize the impact on the folding and structure of target proteins, In addition, two tags in series can fully improve protein solubility (
Bourett and Howard, 2011;
Cheng et al., 2017). Rosetta series of expression strains are derived from the BL21 series of host bacteria and contain rare eukaryotic cell codons in
E. coli, increasing the expression level of heterologous proteins in
E. coli. DE3 is lysogenic λDE3, so it carries a chromosome copy of T7 RNA polymerase, indicating that the strain is suitable for pET series vectors and other T7 promoter series vectors. During purification, molecular sieve chromatography is used to purify the protein in a wide range to avoid the problem of target protein being too small and lost during purification (
Chong et al., 2009;
Lu et al., 2015), Finally, the soluble MGG_140950 protein was successfully obtained. This result laid a theoretical foundation for the study of the rice blast fungus
MGG_
14095 gene involved in metabolic pathways and protein structure analysis, and provided a certain experimental basis for the analysis of the pathogenic mechanism of rice blast fungus and the control of rice blast fungus.
3 Materials and Methods
3.1 Main test materials
The prokaryotic expression vector pETM20 was kept by our laboratory; E. coli competent DH5α and Rosetta (DE3) were purchased from Beijing Zhuangmeng Biotechnology Company. DNA agarose gel recovery kit and plasmid extraction kit were purchased from Beijing Zhuangmeng Biotechnology Company; NcoⅠ, XhoⅠ, DNA Marker, T4 DNA ligase, protein molecular quality standards were purchased from Japan TaKaRa company; Ni agarose gel , Ion exchange chromatography column Resource Q, gel filtration chromatography column SuperdexTM75 were purchased from GE.
3.2 MGG_14095 protein bioinformatics analysis
According to the NCBI website published the MGG_14095 gene sequence (XM_003713271.1) and protein sequence (XP_003713319) of Magnaporthe grisea, the online program ProtParam (https://web.expasy.org/ protparam/) analyzes the primary structure and physical and chemical properties of MGG_14095 protein; Program NCBI program (https://blast.ncbi.nlm.nih.gov/) to analyze MGG_14095 protein homology; online program pfam (http://pfam.xfam.org/search/sequence) to analyze protein conserved domains; Online programs TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/) analyze protein transmembrane domains and signal peptides Location.
3.3 MGG_14095 (38-281) gene cloning and construction of pETM20-MGG_14095 (38-281) vector
Using the rice blast fungus cDNA as a template, NcoⅠ and XhoⅠ were selected as restriction sites based on the vector pETM20 multiple cloning site, and a double primer was designed to amplify the MGG_14095 gene except for the transmembrane structure and signal peptide cleavage site, and was renamed MGG_14095 (38-281) Gene (with F1: 5'-catgcatccaactgccggatccct-3', R1: 5'-ggccgccctttgtggcgtt-3' to amplify the target gene fragment with restriction site NcoⅠ; F2: 5'-catccaactgccggatccct-3', R2: 5'-tcgaggccgcctttgtggcgtt-3' Amplify the target gene fragment with restriction site XhoⅠ). Reaction procedure: 95℃ pre-denaturation for 5 min; 95℃ denaturation for 30 s, 55℃ annealing for 30 s, 72℃ extension for 45 s, 30 cycles; 72℃ extension for 10 min, product verified by 1% agarose gel electrophoresis (same below) and cut The target band was recovered, and the products were mixed at the same concentration to perform nucleic acid molecule hybridization. The hybridization conditions were 95℃ for 10 min, 65℃ for 10 min, 37℃ for 10 min, and 25℃ for 10 min. The results of gene cloning were detected by electrophoresis.
PETM20 and the recovered fragments were double-digested with restriction enzymes to recover the digested products, and ligated with T4 ligase at 16℃ overnight. The heat shock method (
Li, 2018) was used to transform
E. coli DH5α and applied to LB (containing Amp100ug/mL, the same below). Incubate at 37℃ overnight on the plate, pick a single colony for PCR verification, expand the culture by detecting the correct strain and send it to Shanghai Shengong Biological Company for nucleic acid sequence analysis.
3.4 Induction expression and solubility analysis of fusion protein TrxA-MGG_14095 (38-281)
The plasmid extracted the correct expression vector after sequence analysis was renamed to TrxA-MGG_14095 (38-281), transformed into the expression strain Rosetta (DE3) by heat shock method, spread on LB plate and incubated at 37℃ overnight, pick fresh The transformants were cultured in 2 mL liquid LB medium at 37℃ and shaking at 220 r/min until the OD600 was 0.5~0.6. Add 0.1 mmol/L IPTG and continue culturing for 5 h (set the blank control of the inducer). After centrifugation at 12 000 r/min for 5 minutes, the supernatant was collected, 50 μL of 1 Loading Buffer was added to suspend the cells, and the cells were lysed in a boiling water bath for 10 minutes. The induction was detected by SDS-PAGE electrophoresis using 12% separation gel (the same below).
The successfully expressed strains were selected, inoculated into 1 L LB medium at 1:100, and cultured at 37℃ and 220 r/min to achieve an OD600 of about 0.6. IPTG with a final concentration of 0.1 mmol/L was added and induced at 18℃ for 20 h at a low speed. Centrifuge at 5 000 g for 10 min to collect the cells, resuspend the cells using buffer (20 mmol/L Tris-HCl, 500 mmol/L NaCl, pH 8.0), add 1 mmol/L DTT and sonicate for 20 min, then centrifuge at 20℃ at 4℃ At 30 min, take a small amount of supernatant and precipitate for SDS-PAGE detection.
3.5 Fusion protein TrxA-MGG_14095 (38-281) affinity chromatography
Ni-NTA (General Electric Company; American GE company) pretreatment: first treat 2 column volumes with 0.5 mol/L EDTA solution; rinse 3 column volumes with ultrapure water; add 50 mmol/L nickel chloride solution to treat 2 Column volume; Equilibrate 2 column volumes with protein buffer (20 mmol/L Tris-HCl, 500 mmol/L NaCl, 0.5% glycerol, pH 8.0), pour protein lysate into the chromatography column, and collect the eluate. The target protein was eluted with 0 mmol/L, 40 mmol/L, 60 mmol/L, 80 mmol/L, 100 mmol/L, 200 mmol/L, 300 mmol/L, 400 mmol/L, 500 mmol/L imidazole, and the elution result was subjected to SDS-PAGE Detection.
3.6 Fusion protein TrxA-MGG_14095 (38-281) ion exchange chromatography
Based on the fusion protein TrxA-MGG_14095 (38-281) isoelectric point (pI) to select the buffer and buffer pH range, using a 1 mL ion exchange chromatography column as a pre-packed column Resource Q: equilibrated with 5~10 mL low salt buffer Post-affinity desalting treatment The target protein after affinity chromatography (degraded to low or no salt according to TrxA-MGG_14095 protein stability), the concentrated sample is injected into the protein purifier; after washing the chromatography column with low salt buffer , Use low-salt and high-salt buffers (20 mmol/L Tris-HCl, 100 mmol/L NaCl, 0.5% glycerol, pH 8.0) to linearly elute TrxA-MGG_14095 protein at a flow rate of 1 mL/min; collected by SDS-PAGE detection Each peak corresponds to a protein sample.
3.7 Gel filtration chromatography of fusion protein TrxA-MGG_14095 (38-281)
The molecular screening chromatography column used was SupredexTM 75 10/300 GL, the column volume was 24 mL: the protein solution obtained in the previous step was concentrated again to 0.5 mL, and the sample was loaded after equilibrating the chromatography column. Column to complete the elution; peak collection, each tube volume is 1 mL; SDS-PAGE detection of protein samples corresponding to each peak collected.
Authors’ contributions
Wang Shuang is the executor of experimental research, completing data analysis and writing the first draft of the thesis; Li Guorui is involved in the experimental design and analysis of the experimental results; Huang Fenglan and Chen Yongsheng are the project planners and person in charge, guiding the experimental design, data analysis, writing and revision . All authors read and agreed to the final text.
Acknowledgements
This research was jointly funded by the scientific research projects of Inner Mongolia University for Nationalities (MDK2016008, MDK2018018, MDK2019020) and the scientific research projects of Inner Mongolia Autonomous Region colleges and universities (NJZY20121).
Ali O.A., Mvnir T., 2017, Purification and Characterization of Cutinase from Bacillus sp. KY0701 Isolated from Plastic Wastes, Preparative Biochemistry & Biotechnology, 47(9): 925-933
PMid:28857676
Bourett T.M., Howard R.J., 2011, In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea, Canadian Journal of Botany, 68(2): 329-342
Chen X., Shi J., Chen R., Wen Y., Li L., 2015, Molecular chaperones (TrxA, SUMO, Intein and GST) mediating expression, purification and antimicrobial activity assays of plectasin in escherichia coli. Biotechnology and Applied Biochemistry, 62(5): 606-639
PMid:25311837
Cheng Z.X., Ren G.W., Huang J.Y., 2017, Synthesis of Affinity Ionic Liquids for the Purification of Hexahistidine-tagged Proteins, Zhongguo Shipin Xuebao(Journal of Chinese Institute of Food Science and Technology), 17(12): 122-128
Chong F.C., Tan W.S., Biak D.R.A., Ling T.C., Tey B.T., 2009, Purification of histidine-tagged nucleocapsid protein of nipah virus using immobilized metal affinity chromatography, Journal of Chromatography B Analytical Technologies in the Biomedical & Life Sciences, 877(14-15), 1561-1567
PMid:19395325
Deng Y., Zhai K., Xie Z., Yang D., Zhu X., Liu J., Wang X., Qin P., Yang Y., Zhang G., Li Q., Zhang J., Wu S., Milazzo J., Mao B., Wang E., Xie H., Tharreau D., He Z., 2017, Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance, Science, 355(6328): 962-965
PMid:28154240
Duan X., Liu Y., You X., Jiang Z., Yang S., Yang S., 2017, High-level expression and characterization of a novel cutinase from malbranchea cinnamomea suitable for butyl butyrate production. Biotechnology for Biofuels, 10(1), 223
PMid:28932264 PMCid:PMC5606096
Fernandez J., Orth K., 2018, Rise of a Cereal Killer: The Biology of Magnaporthe oryzae Biotrophic Growth, Trends in microbiology, 26(7): 285-597
PMid:29395728 PMCid:PMC6003838
Hamer J.E., Talbot N.J., 1999, Infection-related development in the rice blast fungus Magnaporthe grisea, Current Opinion in Microbiology, 1(6): 693-697
Li X.C., 2018, Castor RcDELLA(GAI) protein expression, purification and crystal growth conditions filtering. Neimenggu Minzu Daxue(Inner Mongolia University For Nationalities)
Lu S.Y., Chen S.X., Yao Y.Y., Xing M.M., Xie Y., 2015, Research of Protein Separation and Purification Methods, Guangzhou Huagong(Guangzhou Chemical Industry), 17: 22-23+37
Martin U.M., Oses R.M., Ryder L.S., Talbot N.J., 2016, Investigating the biology of plant infection by the rice blast fungus magnaporthe oryzae. Fungal Genetics and Biology, 3(90):61-68
PMid:26703899
Qiu F.L., Wang D.W., 2004, Progress in Nosogenesis of the Rice Blast Fungus, Beifang Shuidao(North Rice), 3: 26-28
Roychowdhury M., Jia Y.L., Cartwright R.D., 2013, Structure, Function, and Co-evolution of Rice Blast Resistance Genes, Zuowu Xuebao(Acta Agronomica Sinica)38, 381-393
Ye X., Sun Q., Liu Z., 2015, Progress on Magnaporthe oryzae Infection Process Related to Signaling Pathways, Zhongguo Nongye Keji Daobao (Journal of Agricultural Science and Technology), 17(1): 87-94
Zhang N., Xu D., 2017, Advances of molecular biology techniques in rapid bacterial identification, Guoji Erkexue Zazhi(Progress in Biochemistry and Biophysics), 44(5): 332-335
Zhang X.N., Ran Q.Q., Zhnag X.J., 2013, Research and application progress on cutinase, Zhonguo Niangzao(China Brewing), 32(11): 11-17
 Author
Author  Correspondence author
Correspondence author
.png)
.png)
.png)
.png)
.png)
.png)
.png)

.png)
.png)
.png)


.png)

.png)

.png)



