2. School of Biotechnology, Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Chatha, Jammu, J&K-180009, India
 Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2012, Vol. 3, No. 3 doi: 10.5376/mgg.2012.03.0003
Received: 01 Aug., 2012 Accepted: 07 Aug., 2012 Published: 13 Aug., 2012
Bhavani et al., 2012, Genetic Imprinting in Maize, Maize Genomics and Genetics, Vol.3, No.3, 13-21 (doi: 10.5376/mgg.2012.03.0003)
Genetic imprinting is the epigenetic phenomenon observed in flowering plants including maize in which, an allele shows differential expression depending on the parent it is inherited. Imprinted genes are modified during gametogenesis so that only paternal or maternal allele is expressed after fertilization. This review focuses on the major processes involved in the regulation of imprinting like DNA methylation, covalent modifications of histones and chromatin remodeling citing examples of maize. This review also summarizes theimprinted genes that have been reported in maize including Fie1, Fie2, Peg1, Nrp1, Mez1, Meg1, Mee1, VIP5 and Yuc8.
Transfer of acquired characters of an individual within the lifetime without the change in DNA sequence is termed as “epigenetics” unlike Mendelian genetics, which describes the inheritance of genes that encode specific traits (Ho and Burggren, 2010). Though, the “theory of acquired inheritance” was given by Lamarck way back in 1809 and endorsed by Lysenko (Lysenko, 1948). The concept did not gain much needed attention due to the more convincing evidence obtained by the experiments of Mendel and predecessors (detailed in Weldon, 1902). But inheritances of certain traits like veliger development in snail (Freeman and Lundelins, 1982), epigenetic abberrations and spermatogenesis (Singh et al., 2011), impact of exposure of parents to environmental stress on development of resistance to the environment stress in the progeny (Agrawal et al., 1999) has opened up a new dimension of this, much ignored theory of science. The term “epigenetics” was first coined by Waddington (Waddington, 1942), which literally means “above stress”. Waddington defined epigenetics as “environment-gene interactions that induce developmental phenotype”. But the recent and the most widely accepted definition is ‘‘an epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence” (Singh et al., 2011). The modifications that add this extra piece of information without changing the DNA sequences are DNA methylation, histone modifications and chromatin remodeling proteins. Therefore, the genome is the sum total of the information encoded by the nucleotide sequences while the epigenome is the amassed effect of these DNA and histone modifications on gene expression without affecting the base sequence. Thus, imprinted expression states are under epigenetic control (Springer and Gutierrez-Marcos, 2009). These states can have short-term and long-term effects and could be trans-generational (Anway et al., 2005).
In plants, the evidences of genetic imprinting have been observed in triploid endosperm and not in vegetative tissues (Gehring et al., 2004; Gehring et al., 2009; Huh et al., 2008; Jullien and Berger, 2009; Springer and Gutierrez-Marcos, 2009). Since the active and silenced state of gene is present in the same nucleus, there is the possibility of existence of distinct mechanisms to maintain the epigenetic state between the parental alleles. Maize (Zea mays L.) provides an excellent model to study the role of epigenetic variation and parent-of-origin effect (Eichten et al., 2011; Waters et al., 2011) owing to its large endosperm. Genetically, maize is a highly diverse species (Buckler et al., 2006; Messing and Dooner, 2006) and has complex genome organization with many interspersed genic and repetitive regions (Rabinowicz and Bennetzen, 2006; Schnable et al., 2009). Imprinted genes are arranged in singletons in Arabidopsis and Rice (Gehring et al., 2011; Luo et al., 2011; Wolff et al., 2011) but gene clusters are relatively more frequent in maize (Zhang et al., 2011), in contrast, in mammals, they are found in clusters (Kinoshita, 2004; Gutierrez-Marcos et al., 2006; Jullien, 2006; Feil and Berger, 2007). Imprinted gene expression is classified as paternal imprinting (PEG) or maternal imprinting (MEG) depending upon which parental allele is expressed, allele-specific imprinting (certain alleles at a given locus are imprinted) or gene-specific imprinting (all the alleles at a given locus are imprinted), binary imprinting (monoallelic expression where only one of the parental allele is expressed and other is silent) or differential imprinting (biallelic expression where both alleles are expressed but in different quantities) and constitutive- and transient-imprinted genes depending on the duration of imprinting (Springer and Gutierrez-Marcos, 2009). Imprinting was first reported for R locus in maize which is responsible for aleurone pigmentation of the maize kernel, such as R-r: standard (R-r:std) wherein maize produced fully colored kernel when inherited from female and mottled kernels when paternally inhererited (Kermicle, 1970; Kermicle, 1978; Kermicle and Alleman, 1978; MacDonald, 2012). Although genetic imprinting was first discovered in maize, so far there are only few gene-specific imprinted genes reported in maize (including Fie1, Fie2, Peg1, Nrp1, Mez1, Meg1, and Mee1), most of which are preferentially expressed in the endosperm. All except Peg1 show maternal-specific expression (Zhang et al., 2011). Recent advances in transcriptome profiling techniques like deep sequencing have reported many PEGs and MEGs in Maize endosperm (Waters et al., 2011).
1 Epigenetic mechanisms
Chromatin level of genome activity is controlled at various levels of DNA and histone modifications (Roudier et al., 2011). Covalent modifications of histones, DNA methylation, incorporation of histone variants, and other factors, such as chromatin-remodelling enzymes or small RNAs, all contribute to defining distinct chromatin states that modulate access to DNA (Berger, 2007; Kouzarides, 2007; Roudier et al., 2011). The different epigenetic mechanisms include: a. Modification at the DNA level (Cytosine methylation); b. Modifications at protein level - the histone code (Histone acetylation; Histone methylation; Histone phosphorylation; Histone ubiquitination; Different types of histones); c. Chromatin remodeling - chromatin remodeling proteins.
2 DNA methylation
Methylation patterns of the cytosine residues in the CpG islands serve as one of the important source code in regulating gene expression in epigenetic mechanism. CpG island is a stretch of DNA sequence with high frequency of CpG occurrence and C + G content of more than 50% (Gardiner-Garden and Frommer, 1987; Takai and Jones, 2002) and most commonly observed near promoter regions (Bird et al., 1995). Hypermethylation of DNA in CpG islands is associated with the maintenance of gene suppression, while hypomethylation in these regions is associated with gene expression (Biermann and Steger, 2007). DNA methylation is regulated by DNA methyltransferases that transfer methyl groups from S-adenosyl-methionine to 5' position of cytosine residues of CpG island (Biermann and Steger, 2007). In plants, DNA methylation occurs at cytosine residues in CG, CHG and CHH different sequence contexts (Law and Jacobsen, 2010). Unlike DNMT1 which is involved in maintaining established methylation patterns and DNMT3A and DNMT3B in de novo methylation patterns in mammals (Bestor et al., 1988; Lei et al., 1996; Okano et al., 1999; Singh et al., 2011). Maintenance is carried out by DNA methyltransferase 1 (MET1), variant in methylation (VIM) and decreased DNA methylation 1 (DDM1) (at CG sites), chromomethylase 3 (CMT3) (CHG and CHH) and to some extent de novo CHG methylation is established by domains rearranged methyltransferase 2 (DRM2) in plants (Law and Jacobsen, 2010; Wollmann and Berger, 2012). Twenty-four nucleotide long (24 nt) small interfering RNAs (siRNAs) have shown to mediate De novo DNA methylation through the RNA-directed DNA methylation (RdDM) pathway (Simon and Meyers, 2010). The DNA glycosylase DEMETER (DME) actively removes DNA methylation (Choi et al., 2002; Kinoshita et al., 2004) and might contribute to the derepression of genes (Wollmann and Berger, 2012). DNA demethylation can also occur passively in the absence of enzymes involved in methylation maintenance process (Hauser et al., 2011). In plant endosperm and mammalian embryo, many differentially methylated regions (DMR) are present in the Imprint Control Regions (ICR) that has critical role in epigenetic regulation of imprinted domains (MacDonald, 2011). The methylation pattern of these DMRs are erased in germline, re-established during gametogenesis and maintained throughout the development and lifecycle. Further, DNA methylation is coordinated by the position and composition of nucleosomes and associated histone modifications at genome level (Hauser et al., 2011).
3 Histone modification
The chromatin is made up of nucleosome unit, which is composed of 146 bp and wrapped around an octamer of core histones (H2A, H2B, H3 and H4) and linked by H1. The chemical modifications of the amino acid residue present in the N-terminal tail of these histones result in the regulation of the genes. The modifications viz., methylation, acetylation, phosphorylation, ubiquitylation and sumoylation at the histone tails constitute histone code (Peterson and Laniel, 2004). There are eight common histone modifications that are associated with active or repressed transcriptional state of chromatin (Huan and Springer, 2008). Modifications of histones H3 and H4, especially acetylation and methylation of histone lysine residues at N-terminal tails that protrude from the nucleosome are best understood in terms of gene regulation (Hauser et al., 2011). Histone methylation is the most prominent of the post-translational modification and is monitored by the histone methyltransferases (HMTs). HMTs are involved in either addition or deletion of one or two methyl groups from arginine and lysine residues (Singh et al., 2011). Histone methylation is most commonly associated with the gene silencing, methylation of H3K9 is found in heterochromatin and silenced promoters (Fischle et al., 2003), but it may also associate with gene activation as in histone H3K4 dimethylation in maternal allele of maize Mez1 and ZmFie1 (Huan and Springer, 2008). Therefore, methylation of Lys9 and Lys27 of histone H3 (H3K9 and H3K27) are linked to heterochromatin and gene silencing, while methylation of Lys4 (H3K4) is linked to transcriptional activity (Grewal and Elgin, 2002; McDonald, 2011). Lysine methyltransferase, G9a, is involved in mono- and dimethylation of H3K9 in euchromatin (Tachibana et al., 2007) while LSD1 and JmjC-domain-containing proteins are methyltransferases with lysine demethylase activity that interact with chromatin remodeling proteins to affect chromatin condensation (Okada et al., 2007).
Acetylation is the second most important posttranslational histone modification that has antagonistic role to DNA methylation. Increased histone acetylation at lysine residues is mediated by histone acetyl transferases (HATs) signifies active genes and deacetylation through histone deacetylases (HDACs) inhibit gene expression (Singh et al., 2011). The mouse Gtl2 DMR of the silent paternal allele is hypoacetylated on H3 and H4, while the active maternal allele carries high levels of acetylation on both histones (Carr et al., 2007). MYST1, a MYST family protein is a acetyl transferase (HAT), which acetylates H3K16 to impact chromatin architecture (Neal et al., 2000) while SIRT1 is a deacetylase (HDAC) that removes acetyl groups from H1, H3 and H4 (Yi and Luo, 2010). Phosphorylation of histones at serine and threonine residues and ubiquitylation of lysine residues are associated with either activation or repression of gene depending on the context. For example, phosphorylation is usually associated with gene activation but gene silencing is seen when the histone variant H2AX is phosphorylated (Fernandez-Capetillo, 2003) and ubiquitylation of histone H2A is linked to gene silencing (Baarends et al., 2003) whereas ubiquitylation of H2B is linked to gene activation (Zhu et al., 2005). Attachment of small ubiquitin-related modifier proteins, termed as sumoylation is yet another process of posttranslational modification that mediate gene silencing by recruiting HDACs and heterochromatin protein 1 (Shiio and Eisenman, 2003). DNA methylation and histone modifications are the two interconnected processes in epigenetic mechanisms that influence each other’s recruitment to the silencing complex to reinforce differential epigenetic states (Tariq and Paszkowski, 2004; Cheung and Lau, 2005).
4 Chromatin remodeling
Chromatin structure is associated with the active/repressed state of a gene which is directly influenced by the DNA methylation, histone modifications and chromatin remodeling proteins. Open state of the chromatin makes DNA accessible to transcriptional machinery and gene expression while gene expression is repressed when the chromatin attains more compact state of heterohromatin (Fransz and Jong, 2002). Histone modifications may impact secondary chromatin structures through nucleosome–DNA or nucleosome–nucleosome interactions and by neutralizing charge in the histone N-terminal tails (Wolffe and Hayes, 1999; Carruthers and Hansen, 2000; Wang et al., 2001; Gilbert et al., 2007). The acetylation of histones corresponds with ‘open’ chromatin and enhanced transcriptional activity (Strahl and Allis, 2000) and acetylated histone tails increase the affinity of chromatin for bromo-domain proteins (e.g. HATs) and promote transcriptional activation (Turner, 2000). Chromatin remodeling is mediated by the alterations in location and structure of nucleosomes by ATP-dependent chromatin remodeling proteins (Narlikar et al., 2002; Singh et al., 2011) (e.g. the SWITCH2 [SWI2]/SUCROSE NON-FERMENTING2 [SNF2] complex) and histone- modifying complexes (e.g. the histone deacetylase complex [HDAC]) (Fransz and Jong, 2002). Further, the repressive complex is maintained by the heterochromatin-associated protein HP1 that is thought to form a repressive complex by binding to methylated H3K9 via its chromodomain and by interacting with SUV39 (Fransz and Jong, 2002). A plant homolog of HP1, LHP1 (LIKE HETEROCHROMATIN PROTEIN1), has been reported in Arabidopsis (Gaudin et al., 2001).
5 Imprinted genes in maize
Genetic imprinting is seen in endosperm of flowering plants. During fertilization one of two sperm cells produced by the male gametophyte through meiotic division fuses with egg cell to form seed, the other sperm cell unites with the two central cell of same genetic constitution giving rise to triploid endosperm (Drews and Yadegari, 2002) that serve to nourish embryo (Figure 1). Parental differences in DNA methylation have been identified for the imprinted maize genes ZmFie1 (Hermon et al., 2007), ZmFie2 (Gutierrez-Marcos et al., 2006), Mez1 (Haun et al., 2007), Peg1 (Gutierrez-Marcos et al., 2003), Nrp1 (Guo et al., 2003; Haun and Springer, 2008), Meg1 (Gutierrez-Marcos et al., 2004), Mee1 (Jahnke and Scholten, 2009) and VIM5 and YUC10 (Zhang et al., 2011) (Table 1). All the reported imprinted genes are preferentially expressed in the endosperm and all except Peg1, VIM5 and YUC10 show maternal-specific expression (Gutierrez-Marcos, 2003). Two types of imprinting exist in maize, allele-specific imprinting as seen in R gene (Kermicle, 1970), dzr1 gene and zein protein (Chaudhuri and Messing, 1994; Lund et al., 1995a), alpha-tubulin (Lund et al., 1995b) and gene-specific imprinting as in ZmFie1 (Danilevskaya et al., 2003) and Nrp1 (Guo et al., 2003). Among the maize genes, few alleles of the R locus such as R-r: standard (R-r:std) was first reported to be imprinted (Kermicle, 1970). This R-r:std allele is responsible for the aleurone pigmentation of maize which shows differential expression depending on the contributing parent (Kermicle, 1970). The paternally inherited R allele gives mottled phenotype while maternally inherited R allele gives solid phenotype (Kermicle, 1970). The imprinting of R allele appears to be due to differential expression of the maternal allele in relation to paternal allele rather than silencing of paternal allele (Kermicle and Alleman, 1990). Alpha-tubulin genes exhibit differential methylation depending on whether it is maternal or paternal inheritance (Lund et al., 1995). Locus dzr1, a posttranscriptional regulator of zein protein also shows allele-specific imprinted expression (Chaudhuri and Messing, 1994).
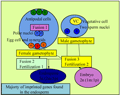 Figure 1 Fertilization events in the angiosperms |
.png) Table 1 Imprinted genes in maize and their expression pattern |
ZmFie1 and ZmFie2 are maize orthologs of the Arabidopsis FIE gene, (Gutierrez-Marcos et al., 2006; Raissig et al., 2011). ZmFie1 is imprinted in the endosperm, which shows maternal expression, and paternal expression is completely absent throughout seed development, which is repressed by DNA methylation of CpG island (Danilevskaya et al., 2003; Gutierrez-Marcos et al., 2006; Huan and Springer; 2008). No-apical meristem (NAM) related protein1 (Nrp1), a transcription factor, displayed gene-specific imprinting and expressed only in endosperm (Guo et al., 2003). Janhnke and Scholten (2009), has shown the imprinting of maternally expressed in embryo 1 (mee1) gene of maize in both the embryo and endosperm indicating the correlation of parent-of-origin-specific expression with differential allelic methylation. This differential methylation is maintained in the endosperm, whereas the embryonic maternal allele is demethylated on fertilization and remethylated later in embryogenesis. Maternally expressed gene1 (meg1), which is a maize endosperm transfer cell-specific gene shows maternal parent-of-origin expression pattern during early stages of endosperm development but biallelic expression at later stages (Gutierrez-Marcos et al., 2004). Polycomb group (PcG) proteins, Maize Enhancer-of-zeste 1 (Mez1) is imprinted while Mez2 and Mez3 are not. Mono-allelic expression of maternal Mez1 was seen in the developing endosperm and was found to be present throughout the development of endosperm. (Huan et al., 2009). Differentially methylated region was found at the transcription start site of Mez1 and CpG and CpNpG nucleotides were heavily methylated on the non-expressed paternal allele while maternal allele showed less methylation (Huan et al., 2007). Recent advances in transcriptome profiling techniques like deep sequencing have reported many PEGs and MEGs in Maize endosperm (Wang et al., 2009). Waters et al (2011), reported 100 putative imprinted genes in maize endosperm, including 54 maternally expressed genes (MEGs) and 46 paternally expressed genes (PEGs) using genome-wide allele-specific expression profiling. Zhang et al showed 179 genes (1.6% of protein-coding genes) expressed in the endosperm to be imprinted, with 68 of them showing maternal preferential expression and 111 paternal preferential expressions. Zhang et al (2011) has identified two PEGs VIM5 and Yuc10 in maize.
6 Conclusion and our views
Genetic imprinting is observed only in the triploid endosperm of the angiosperms in the plant kingdom and around thirteen imprinted genes have been reported in maize. Hundreds of imprinted genes are being added to the list through deep sequencing technologies, but the functional role is yet to be elucidated. DNA methylation and histone modifications are the two important process involved in exertion and maintenance of imprinting. Genetic imprinting is found to have major role in many key developmental processes and genome dosage is one of the factors contributing to the imprinting. Genome dosage has reported to have direct implication on the seed size in maize. Though we can speculate that the role of imprinting in the maintenance of the genome dosage imbalance in the cross-pollinated plants, its role in self-pollinated plants is yet to be discerned. To date, the scientific community is still debating on its role in evolution and significance in the process of crop improvement. Advanced technologies like genome-wide approaches may contribute in helping the researchers to unravel the potential mechanism of genetic imprinting and its possible benefits to crop improvement.
References
Agrawal A.A., Laforsch C., and Tollrian R., 1999, Transgenerational induction of defences in animals and plants, Nature, 401: 60-63
http://dx.doi.org/10.1038/43425
Anway M.D., Cupp A.S., Uzumcu M., Skinner M.K., 2005, Epigenetic transgenerational actions of endocrine disruptors and male fertility, Science, 308: 1466-1469
http://dx.doi.org/10.1126/science.1108190 PMid:15933200
Baarends W.M., Wassenaar E., Hoogerbrugge J.W., van Cappellen G., Roest H.P., Vreeburg J., Ooms M., Hoeijmakers J.H., and Grootegoed J.A., 2003, Loss of HR6B ubiquitin- conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase, Mol. Cell Biol., 23: 1151-1162
http://dx.doi.org/10.1128/MCB.23.4.1151-1162.2003 PMid:12556476 PMCid:141135
Baarends W.M., Wassenaar E., Hoogerbrugge J.W., van Cappellen G., Roest H.P., Vreeburg J., Ooms M., Hoeijmakers J.H., Grootegoed J.A., 2003, Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase, Mol. Cell Biol., 23: 1151-1162
http://dx.doi.org/10.1128/MCB.23.4.1151-1162.2003 PMid:12556476 PMCid:141135
Berger S.L., 2007, The complex language of chromatin regulation during transcription, Nature, 447: 407-412
http://dx.doi.org/10.1038/nature05915 PMid:17522673
Bestor T., Laudano A., Mattaliano R., and Ingram V., 1988, Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases, J. Mol. Biol., 203: 971-983
http://dx.doi.org/10.1016/0022-2836(88)90122-2
Biermann K., and Steger K., 2007, Epigenetics in male germ cells, J. Androl., 28: 466-480
http://dx.doi.org/10.2164/jandrol.106.002048 PMid:17287457
Bird A., Tate P., Nan X., Campoy J., Meehan R., Cross S., Tweedie S., Charlton J., and Macleod, D., 1995, Studies of DNA methylation in animals, J. Cell Sci. Suppl., 19: 37-39
PMid:8655645
Buckler E.S., Gaut B.S., and McMullen M.D., 2006, Molecular and functional diversity of maize, Curr. Opin. Plant Biol., 9: 172-176
http://dx.doi.org/10.1016/j.pbi.2006.01.013 PMid:16459128
Carr M.S., Yevtodiyenko A., Schmidt C.L., and Schmidt J.V., 2007, Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain, Genomics, 89(2): 280-290
http://dx.doi.org/10.1016/j.ygeno.2006.10.005 PMid:17126526 PMCid:1802099
Carruthers L.M., and Hansen J.C., 2000, The core histone N termini function independently of linker histones during chromatin condensation, J. Biol. Chem., 275: 37285-37290
http://dx.doi.org/10.1074/jbc.M006801200 PMid:10970897
Chaudhuri S., and Messing J., 1994, Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation, Proc. Natl. Acad. Sci., 91: 4867-4871
http://dx.doi.org/10.1073/pnas.91.11.4867
Cheung P., and Lau P., 2005, Epigenetic regulation by histone methylation and histone variants, Molecular Endocrinology, 19(3): 563-573
http://dx.doi.org/10.1210/me.2004-0496 PMid:15677708
Choi Y., Gehring M., Johnson L., Hannon M., Harada J.J., Goldberg RB., Jacobsen S.E., and Fischer R.L., 2002, DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis, Cell, 110: 33-42
http://dx.doi.org/10.1016/S0092-8674(02)00807-3
Danilevskaya O.N., Hermon, P., Hantke S., Muszynski M.G., Kollipara K., and Ananiev E.V., 2003, Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions, The Plant Cell, 15: 425-438
http://dx.doi.org/10.1105/tpc.006759 PMCid:141211
Drews G.N., and Yadegari R., 2002, Development and function of the angiosperm female gametophyte, Annu. Rev. Genet., 36: 99-124
http://dx.doi.org/10.1146/annurev.genet.36.040102.131941 PMid:12429688
Eichten S.R., Swanson-Wagner R.A., Schnable J.C., Waters A.J., Hermanson P.J., Liu S., Yeh C.T., Jia Y., Gendler K., Freeling M., Schnable P.S., Vaughn M.W., and Springer N.M., 2011, Heritable epigenetic variation among maize inbreds. PLoS Genet., 7: e1002372
http://dx.doi.org/10.1371/journal.pgen.1002372 PMid:22125494 PMCid:3219600
Feil R., and Berger F., 2007, Convergent evolution of genomic imprinting in plants and mammals, Trends Genet., 23: 192-199
http://dx.doi.org/10.1016/j.tig.2007.02.004 PMid:17316885
Fernandez-Capetillo O., Mahadevaiah S.K., Celeste A., Romanienko P.J., Camerini-Otero R.D., Bonner W.M., Manova K., Burgoyne P., and Nussenzweig A., 2003, H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis, Dev. Cell, 4: 497-508
http://dx.doi.org/10.1016/S1534-5807(03)00093-5
Fischle W., Wang Y., Jacobs S.A., Kim Y., Allis C.D., and Khorasanizadeh S., 2003, Molecular basis for the discrimination of repressive methyllysine marks in histone H3 by polycomb and HP1 chromodomains, Genes Dev., 17: 1870-1881
http://dx.doi.org/10.1101/gad.1110503
Fransz F.P., and Jong J.H., 2002, Chromatin dynamics in plants, Current Opinion in Plant Biology, 5: 560-567
http://dx.doi.org/10.1016/S1369-5266(02)00298-4
Freeman G., and Lundelius J.W., 1982, The developmental genetics of dextrality and sinistrality in the gastropod Lymnaea peregra, Roux Arch. Dev. Biol., 191: 69-83
http://dx.doi.org/10.1007/BF00848443
Gardiner-Garden M., and Frommer M., 1987, CpG islands in vertebrate genomes, J. Mol. Biol., 2: 261-282
http://dx.doi.org/10.1016/0022-2836(87)90689-9
Gaudin V., Libault M., Pouteau S., Juul T., Zhao G., Lefebvre D., and Grandjean O., 2001, Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis, Development, 128: 4847-4858
PMid:11731464
Gehring M., Bubb K.L., and Henikoff S., 2009, Extensive demethylation of repetitive elements during seed development underlies gene imprinting, Science, 324(14): 47-51
PMid:19342577
Gehring M., Choi Y., and Fischer R.L., 2004, Imprinting and seed development, The Plant Cell, 16(Sl): 203-213
Gehring M., Missirian V., and Henikoff S., 2011, Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds, PLoS One, 6: e23687
http://dx.doi.org/10.1371/journal.pone.0023687 PMid:21858209 PMCid:3157454
Grewal S.I., Elgin S.C., 2002, Heterochromatin: new possibilities for the inheritance of structure, Curr. Opin. Genet. Dev., 12: 178-87
http://dx.doi.org/10.1016/S0959-437X(02)00284-8
Guo M., Rupe M.A., Danilevskaya O.N., Yang X., and Hu Z., 2003, Genome-wide mRNA proï¬ling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm, Plant J., 36: 30-44
http://dx.doi.org/10.1046/j.1365-313X.2003.01852.x PMid:12974809
Gutierrez-Marcos J.F., Costa L.M., Biderre-Petit C., Khbaya B., O’Sullivan D.M., Wormald M., Perez P., and Dickinson H.G., 2004, Maternally expressed gene 1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression, Plant Cell, 16: 1288-1301
http://dx.doi.org/10.1105/tpc.019778 PMid:15105441 PMCid:423216
Gutierrez-Marcos J.F., Costa L.M., Dal Pra M., Scholten S., Kranz E., Perez P., and Dickinson H.G., 2006, Epigenetic asymmetry of imprinted genes in plant gametes, Nat. Genet., 38: 876-878
http://dx.doi.org/10.1038/ng1828 PMid:16823380
Gutierrez-Marcos J.F., Pennington P.D., Costa L.M., and Dickinson H.G., 2003, Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci., 358: 1105-1111
http://dx.doi.org/10.1098/rstb.2003.1292 PMid:12831476 PMCid:1693205
Haun W.J., and Springer N.M., 2008, Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes, Plant J., 56: 903-912
http://dx.doi.org/10.1111/j.1365-313X.2008.03649.x PMid:18694457
Haun W.J., Laoueillé-Duprat S., O’connell M.J., Spillane C., Grossniklaus U., Phillips A.R., Kaeppler S.M., and Springer N.M., 2007, Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs, Plant J., 49: 325-37
http://dx.doi.org/10.1111/j.1365-313X.2006.02965.x PMid:17181776
Hauser M., Aufsatz W., Jonak C., Luschnig C., 2011, Transgenerational epigenetic inheritance in plants, Biochim. Biophys. Acta, doi:10.1016/j.bbagrm.2011.03.007
http://dx.doi.org/10.1016/j.bbagrm.2011.03.007 PMid:21515434
Hermon P., Srilunchang K.O., Zou J., Dresselhaus T., Danilevskaya O.N., 2007, Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele, Plant Mol. Biol., 64: 387-95
http://dx.doi.org/10.1007/s11103-007-9160-0 PMid:17437065
Ho D.H., and Burggren W.W., 2010, Epigenetics and transgenerational transfer: a physiological perspective, J. Expt. Biology, 213: 3-16
http://dx.doi.org/10.1242/jeb.019752 PMid:20008356
Huh J.H., Bauer M.J., Hsieh T.F., and Fischer R.L., 2008, Cellular programming of plant gene imprinting, Cell, 132: 735-744
http://dx.doi.org/10.1016/j.cell.2008.02.018 PMid:18329361
Jahnke S., and Scholten S., 2009, Epigenetic resetting of a gene imprinted in plant embryos, Curr. Biol., 19: 1677-1681
http://dx.doi.org/10.1016/j.cub.2009.08.053 PMid:19781944
Jullien P.E., 2006, Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting, The Plant Cell, 18: 1360-1372
http://dx.doi.org/10.1105/tpc.106.041178 PMid:16648367 PMCid:1475502
Jullien P.E., and Berger F., 2009, Gamete-specific epigenetic mechanisms shape genomic imprinting, Curr. Opin. Plant Biol., 12: 637-642
http://dx.doi.org/10.1016/j.pbi.2009.07.004 PMid:19709923
Kermicle J.L., 1978, Imprinting of gene action in maize endosperm. In: Walden D.B. (ed.), Maize Breeding and Genetics, Wiley, New York, pp.357-371
Kermicle J.L., and Alleman M., 1990, Gametic imprinting in maize in relation to the angiosperm life cycle, Development, Suppl.: 9-14
PMid:2090436
Kermicle, J.L., 1970, Dependence of the R-mottle aleurone phenotype in maize on mode of sexual transmission, Genetics, 66: 69-85
PMid:17248508 PMCid:1212486
Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S.E., Fischer R.L., and Kakutani T., 2004, One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation, Science, 303: 521-523
http://dx.doi.org/10.1126/science.1089835 PMid:14631047
Kouzarides T., 2007, Chromatin modifications and their function, Cell, 128: 693-705
http://dx.doi.org/10.1016/j.cell.2007.02.005 PMid:17320507
Law J.A., and Jacobsen S.E., 2010, Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat. Rev. Genet., 11: 204-220
http://dx.doi.org/10.1038/nrg2719 PMid:20142834 PMCid:3034103
Lei H., Oh S.P., Okano M., Juttermann R., Goss K.A., Jaenisch R., and Li E., 1996, De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells, Development, 122: 3195-3205
PMid:8898232
Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R., 1989, Lc, a member of the maize R gene family responsible fortissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci., 86: 7092-7096
http://dx.doi.org/10.1073/pnas.86.18.7092
Lund G., Ciceri P., and Viotti A., 1995a, Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L., Plant J., 8: 571-581
http://dx.doi.org/10.1046/j.1365-313X.1995.8040571.x PMid:7496402
Lund G., Messing J., and Viotti A., 1995b., Endosperm-specific demethylation and activation of specific alleles of alpha-tubulin genes of Zea mays L., Mol. Gen. Genet., 246: 716-722
http://dx.doi.org/10.1007/BF00290717
Luo M., Taylor J.M., Spriggs A., Zhang H., Wu X., and Russell S., 2011, A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm, PLoS Genet., 7: e1002125
http://dx.doi.org/10.1371/journal.pgen.1002125 PMid:21731498 PMCid:3121744
Lysenko T., 1948, The science of biology today, New York: International Publishers
MacDonald W.A., 2011, Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects, Genetics Research International, 2012, Article ID 585024
Messing J., and Dooner H.K., 2006, Organization and variability of the maize genome, Curr. Opin. Plant Biol., 9: 157-163
http://dx.doi.org/10.1016/j.pbi.2006.01.009 PMid:16459130
Narlikar G.J., Fan H.Y., and Kingston R.E., 2002, Cooperation between complexes that regulate chromatin structure and transcription, Cell, 108: 475-487
http://dx.doi.org/10.1016/S0092-8674(02)00654-2
Neal K.C., Pannuti A., Smith E.R., and Lucchesi J.C., 2000, A new human member of the MYST family of histone acetyl transferases with high sequence similarity to drosophila MOF, Biochim. Biophys. Acta, 1490: 170-174
http://dx.doi.org/10.1016/S0167-4781(99)00211-0
Nick G., Thomson I., Boyle S., Allan J., Ramsahoye B., and Bickmore W.A., 2007, DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction, Journal of Cell Biology, 177(3): 401-411
http://dx.doi.org/10.1083/jcb.200607133 PMid:17485486 PMCid:2064831
Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y., 2007, Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis, Nature, 450: 119-123
http://dx.doi.org/10.1038/nature06236 PMid:17943087
Okano M., Bell D.W., Haber D.A., and Li E., 1999, DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development, Cell, 99: 247-257
http://dx.doi.org/10.1016/S0092-8674(00)81656-6
Peterson C.L., and Laniel M.A., 2004, Histones and histone modifications, Curr. Biol., 14(14): R546-R551
http://dx.doi.org/10.1016/j.cub.2004.07.007 PMid:15268870
Rabinowicz P.D., Bennetzen J.L., 2006, The maize genome as a model for efficient sequence analysis of large plant genomes, Curr. Opin. Plant Biol., 9: 149-156
http://dx.doi.org/10.1016/j.pbi.2006.01.015 PMid:16459129
Raissig M.T., Baroux C., and Grossniklaus U., 2011, Regulation and flexibility of genomic imprinting during seed development, The Plant Cell 23: 16-26
http://dx.doi.org/10.1105/tpc.110.081018 PMid:21278124 PMCid:3051244
Roudier F., Ahmed I., Berard C., Sarazin A., Mary-Huard T., Cortijo S., Bouyer D., Caillieux E., Duvernois-Berthet E., Al-Shikhley L., Giraut L., Despres B., Drevensek S., Barneche F., Derozier S., Brunaud V., Aubourg S., Schnittger A., Bowler C., Martin-Magniette M., Robin S., Caboche M., and Colot V., 2011, Integrative epigenomic mapping defines four main chromatin states in Arabidopsis, EMBO J., 2011: 1-11
Schnable P.S., Ware D., Fulton R.S., Stein J.C., and Wei F., 2009, The B73 maize genome: Complexity, diversity, and dynamics, Science, 326 (5956): 1112-1115
http://dx.doi.org/10.1126/science.1178534 PMid:19965430
Shiio Y., and Eisenman R.N., 2003, Histone sumoylation is associated with transcriptional repression, Proc. Natl. Acad. Sci., 100: 13225-13230
http://dx.doi.org/10.1073/pnas.1735528100 PMid:14578449 PMCid:263760
Simon S.A., and Meyers B.C., 2010, Small RNA-mediated epigenetic modifications in plants, Curr. Opin. Plant Biol., 14: 148-155
http://dx.doi.org/10.1016/j.pbi.2010.11.007 PMid:21159545
Singh R., Avery K., and Agarwal A., 2011, Epigenetics, spermatogenesis and male infertility, Mutation Research, 727: 62-71
http://dx.doi.org/10.1016/j.mrrev.2011.04.002 PMid:21540125
Springer N.M., and Gutierrez-Marcos J.F., 2009, Imprinting in maize, Maize Handbook-volume II: Genetics and Genomics, Bennetzen J.L., and Hake S. (eds.), 428-440
Strahl B.D., and Allis C.D., 2000, The language of covalent histone modifications, Nature, 403: 41-45
http://dx.doi.org/10.1038/47412 PMid:10638745
Tachibana M., Matsumura Y., Fukuda M., Kimura H, and Shinkai Y., 2008, G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription, EMBO J., 27, 2681-2690
http://dx.doi.org/10.1038/emboj.2008.192 PMid:18818694 PMCid:2572175
Tachibana M., Nozaki M., Takeda N., and Shinkai Y., 2007, Functional dynamics of H3K9 methylation during meiotic prophase progression, EMBO J., 26: 3346-3359
http://dx.doi.org/10.1038/sj.emboj.7601767 PMid:17599069 PMCid:1933398
Takai D., and Jones P.A., 2002, Comprehensive analysis of CpG islands in human chromosomes 21 and 22, Proc. Natl. Acad. Sci., 99: 3740-3745
http://dx.doi.org/10.1073/pnas.052410099 PMid:11891299 PMCid:122594
Tariq M., and Paszkowski J., 2004, DNA and histone methylation in plants, Trends in Genetics, 20(6): 244-251
http://dx.doi.org/10.1016/j.tig.2004.04.005 PMid:15145577
Turner B.M., 2000, Histone acetylation and an epigenetic code, Bioessays, 22: 836-845
http://dx.doi.org/10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X
http://dx.doi.org/10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.3.CO;2-O
Waddington C., 1942, The epigenotype, Endeavour, 1: 18-20
Wang X., He C., Moore S.C., and Ausio J., 2001, Effects of histone acetylation on the solubility and folding of the chromatin fiber, J. Biol. Chem., 276: 12764-12768
http://dx.doi.org/10.1074/jbc.M100501200 PMid:11279082
Wang Z., Gerstein M., and Snyder M., 2009, RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet., 10(1): 57-63
http://dx.doi.org/10.1038/nrg2484 PMid:19015660 PMCid:2949280
Waters A.J., Makarevitch I., Eichten S.R., Swanson-Wagner R.A., Yeh C., Xu W., Schnable P.S., Vaughn M.W., Gehring M., and Springer N.M., 2011, Parent-of-Origin effects on gene expression and DNA methylation in the maize endosperm, The Plant Cell, 23: 4221-4233
http://dx.doi.org/10.1105/tpc.111.092668 PMid:22198147
Weldon W.F.R., 1902, Mendel's laws of alternative inheritance in peas, Biometrika, 1: 228-254
http://dx.doi.org/10.1093/biomet/1.2.228
Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M.T., 2011, High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm, PLoS Genet, 7: e1002126
http://dx.doi.org/10.1371/journal.pgen.1002126 PMid:21698132 PMCid:3116908
Wolffe A.P., and Hayes J.J., 1999, Chromatin disruption and modification, Nucleic Acids Res., 27: 711-720
http://dx.doi.org/10.1093/nar/27.3.711 PMid:9889264 PMCid:148238
Wollmann H., and Berger F., 2012, Epigenetic reprogramming during plant reproduction and seed development, Current Opinion in Plant Biology, 15: 63-69
http://dx.doi.org/10.1016/j.pbi.2011.10.001 PMid:22035873
Yi J., and Luo J., 2010, SIRT1 and p53, effect on cancer, senescence and beyond, Biochim. Biophys. Acta, 1804: 1684-1689
http://dx.doi.org/10.1016/j.bbapap.2010.05.002
Zhang M., Zhao H., Xie S., Chen J., Xu Y., Wang K., Zhao H., Guan H., Hu X., Jiao Y., Song W., and La J., 2011, Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm, Proc. Natl. Acad. Sci., 108: 20042-20047
http://dx.doi.org/10.1073/pnas.1112186108 PMid:22114195 PMCid:3250141
Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D., 2005, Mono-ubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation, Molecular Cell, 20: 601-611
http://dx.doi.org/10.1016/j.molcel.2005.09.025 PMid:16307923
. PDF(690KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Bhavani P.
. Harinikumar K.M.
. Shashidhar H.E.
. Sajad Majeed Zargar
Related articles
. Maize
. Genetic imprinting
. Epigenetics
. Histone methylation
Tools
. Email to a friend
. Post a comment


