 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2024, Vol. 15, No. 5 doi: 10.5376/tgg.2024.15.0024
Received: 18 Aug., 2024 Accepted: 20 Sep., 2024 Published: 02 Oct., 2024
Zhou J., and Liu X.M., 2024, Wheat epigenetics and its role in crop improvement, Triticeae Genomics and Genetics, 15(5): 255-265 (doi: 10.5376/tgg.2024.15.0024)
Epigenetics, as a mechanism for regulating gene expression, is showing immense potential in agriculture, particularly in the improvement of wheat traits. This study explores the application and potential of epigenetics in wheat crop improvement, reviewing recent progress in wheat epigenetics research. It provides an in-depth analysis of how epigenetic regulation influences wheat yield, disease resistance, and environmental adaptability. Additionally, it summarizes how epigenetic variation can transmit traits through non-genetic inheritance and discusses the prospects of integrating modern breeding technologies such as gene editing. The study highlights the significant role of epigenetics in wheat crop improvement, suggesting that the future of wheat breeding will become more efficient and precise through the application of epigenetic markers, utilization of stable non-genetic variation, and epigenetic plasticity enhancement. The integration of epigenetic techniques with traditional breeding methods, especially with gene editing, is expected to drive the development of precision wheat breeding, addressing the challenges of global food production. Research in this field not only contributes to enhancing wheat stress resilience but also provides new strategies for breeding in the context of global climate change.
1 Introduction
Wheat (Triticum aestivum) is one of the most important staple food crops globally, contributing significantly to the dietary calories and proteins consumed by humans. It plays a crucial role in global food security, providing about 20% of the total dietary calories and proteins worldwide. The crop's importance is underscored by its extensive cultivation, with modern wheat varieties being rapidly adopted across developing regions, accounting for roughly 53% of the total harvested area and 50% of the production (Shiferaw et al., 2013). Wheat's versatility and nutritional value, including its high content of dietary fiber, vitamins, and essential minerals, make it indispensable in both temperate zones and countries undergoing urbanization and industrialization (Shewry and Hey, 2015).
Epigenetics, the study of heritable changes in gene expression that do not involve changes to the underlying DNA sequence, has emerged as a pivotal field in crop improvement. Epigenetic mechanisms, such as DNA methylation, histone modification, and RNA interference, play critical roles in regulating plant development, stress responses, and adaptation to environmental changes (Pang et al., 2020; Li et al., 2021). These mechanisms offer promising avenues for enhancing crop resilience, yield, and nutritional quality without the need for genetic modification, thus addressing some of the challenges posed by traditional breeding methods (Shrawat and Armstrong, 2018).
In wheat, the application of epigenetics holds particular promise. The hexaploid nature of wheat, with its complex genome and gene redundancy, presents unique challenges for genetic research and precision breeding (Li et al., 2021). However, epigenetic modifications can potentially bypass these challenges by regulating gene expression in a more targeted and efficient manner. For instance, epigenetic modifications can enhance wheat's resistance to diseases and pests, improve its adaptation to climate change, and increase its nutritional value (Pang et al., 2020; Khalid et al., 2023).
This study will explore the specific applications and potential of epigenetic mechanisms in wheat crop improvement, providing an in-depth analysis of the current status of wheat epigenetics research. It will highlight key findings from recent studies and discuss the future prospects and challenges in this field. The study aims to offer insights into how epigenetic approaches can contribute to the sustainable intensification of wheat production, addressing global food security challenges.
2 Current Status of Wheat Epigenetics Research
2.1 Analysis of wheat epigenome and technological advances
The analysis of the wheat epigenome has made significant strides, particularly with the advent of high-throughput sequencing technologies. These advancements have enabled researchers to overcome the challenges posed by the large and complex allohexaploid genome of wheat. Recent studies have provided comprehensive epigenomic maps that reveal the intricate regulatory networks governing wheat development and stress responses. For instance, a detailed comparison of epigenomes and transcriptomes across various developmental stages and environmental conditions has identified thousands of distal epigenetic regulatory elements (distal-epiREs) linked to their target promoters. This study highlighted the role of subgenome-divergent activity of homologous regulatory elements, influenced by specific epigenetic signatures such as H3K27me3, mediated by the Polycomb complex and demethylases (Wang et al., 2021).
Technological advances have also facilitated the integration of epigenomic information into crop improvement strategies (Huang, 2024). The use of chromatin profiles to enhance the understanding of complex crop genomes has been a focal point. This approach, termed 'epigenome guided' improvement, leverages chromatin information to better annotate and decode plant genomes. It also aims to identify and select heritable epialleles that control crop traits independently of the underlying genotype. The integration of epigenomic data with CRISPR/Cas9 gene editing and precision genome engineering holds promise for future crop improvement endeavors (Zhang et al., 2022).
2.2 Epigenetic regulatory elements in the wheat genome
Epigenetic regulatory elements play a crucial role in the regulation of gene expression in wheat. These elements, including DNA methylation, histone modifications, and non-coding RNAs, contribute to the dynamic regulation of the wheat genome. A comprehensive review of epigenetic mechanisms has highlighted the importance of these modifications in plant responses to biotic and abiotic stresses. DNA methylation, histone post-translational modifications, and RNA-directed DNA methylation create memory marks that help plants survive various stresses through physiological regulation based on their epigenetic history (Samantara et al., 2021).
The coordinated regulation of early meiotic stages in wheat is dominated by non-coding RNAs and stage-specific transcription. A study focusing on the meiotic transcriptome revealed significant enrichment of non-coding RNAs during prophase I, which controlled the reprogramming of central metabolic pathways. This study identified 9,309 meiosis-specific transcripts and many known and novel non-coding RNAs differentially expressed at specific stages, providing new insights into the regulatory controls of meiosis in wheat (Jiang et al., 2023).
2.3 Application of epigenetic tools and techniques
The application of epigenetic tools and techniques in wheat research has opened new avenues for crop improvement. Epigenetic modifications, such as DNA methylation and histone modifications, have been shown to play a role in developmental gene regulation, response to environmental stimuli, and natural variation of gene expression levels. These modifications can be harnessed to select for favorable epigenetic states, create novel epialleles, and regulate transgene expression, thereby contributing to crop improvement strategies (Springer, 2013).
Recent research has focused on exploiting both induced and natural epigenetic variation for crop improvement. Understanding the sources of epigenetic variation and the stability of newly formed epigenetic variants over generations is crucial for fully utilizing the potential of epigenetic variation. The development and application of methods for widespread epigenome profiling and engineering are expected to generate new avenues for using the full potential of epigenetics in crop improvement. This includes the use of epigenetic diversity to predict plant performance and increase final crop production, particularly in response to climate change (Springer and Schmitz, 2017; Kakoulidou et al., 2021).
3 Major Epigenetic Mechanisms in Wheat
3.1 The role of DNA methylation
DNA methylation is a crucial epigenetic mechanism that involves the addition of a methyl group to the cytosine residues in DNA, typically at CpG sites. This modification can lead to the repression of gene expression by altering the chromatin structure, making it less accessible to transcriptional machinery. In wheat, DNA methylation plays a significant role in regulating responses to environmental stresses such as drought, salinity, and pathogen attacks. For instance, methylation patterns can change in response to these stresses, thereby modulating the expression of stress-responsive genes to enhance plant resilience (Agarwal et al., 2020; Kong et al., 2020).
Moreover, DNA methylation is involved in the regulation of developmental processes in wheat, such as flowering time and seed development. These processes are critical for crop yield and quality. By understanding and manipulating DNA methylation patterns, researchers aim to develop wheat varieties with improved traits, such as higher yield and better stress tolerance. This approach, known as epibreeding, leverages epigenetic variations to complement traditional breeding methods, offering a promising avenue for crop improvement (Samantara et al., 2021; Gupta and Salgotra, 2022).
3.2 Functions of histone modifications
Histone modifications, including methylation, acetylation, phosphorylation, and ubiquitination, are post-translational modifications that occur on the histone proteins around which DNA is wrapped. These modifications can either activate or repress gene expression by altering the chromatin structure and recruiting specific regulatory proteins. In wheat, histone modifications are essential for regulating gene expression in response to both biotic and abiotic stresses. For example, histone acetylation is generally associated with gene activation and has been linked to the expression of genes involved in stress responses (Chang et al., 2019; Agarwal et al., 2020).
Histone modifications also play a crucial role in developmental processes such as flowering and seed germination. These modifications can create a "memory" of environmental conditions, allowing wheat plants to better adapt to changing environments. The dynamic nature of histone modifications makes them a versatile tool for fine-tuning gene expression. Recent advances in understanding the cross-talk between different histone modifications have further highlighted their complex regulatory roles, offering new insights into how these modifications can be targeted for crop improvement (Molina-Serrano et al., 2013; Shafiq and Khan, 2015).
3.3 The relationship between wheat chromatin structure and trait expression
The chromatin structure in wheat is a dynamic entity that can be remodeled in response to developmental cues and environmental signals. Chromatin remodeling involves changes in the positioning of nucleosomes, the basic units of chromatin, which can either facilitate or hinder access to the underlying DNA. This process is crucial for the regulation of gene expression and, consequently, for the expression of various traits in wheat. For instance, chromatin remodeling has been shown to play a role in the regulation of genes involved in stress responses, flowering time, and other important agronomic traits (Shafiq and Khan, 2015; Varotto et al., 2020).
The relationship between chromatin structure and trait expression is also influenced by the interplay between DNA methylation and histone modifications. These epigenetic marks can work together to establish and maintain specific chromatin states that are conducive to either gene activation or repression. Understanding this interplay is essential for developing strategies to manipulate chromatin structure for crop improvement. By targeting specific chromatin remodeling factors or modifying epigenetic marks, researchers aim to enhance the expression of desirable traits in wheat, such as increased yield, improved stress tolerance, and better nutritional quality (Saravana Kumar et al., 2020; Samantara et al., 2021).
4 The Influence of Epigenetic Regulation on Agronomic Traits of Wheat
4.1 Regulation of yield traits in wheat
Epigenetic modifications, such as DNA methylation and histone modifications, play a crucial role in regulating yield traits in wheat. These modifications can influence gene expression without altering the underlying DNA sequence, thereby affecting plant growth and development. For instance, epigenetic changes can enhance yield by improving traits such as spike seed-setting and grain size. Studies have shown that targeted epigenetic editing can amplify yield potential by modulating specific genes associated with these traits (Pang et al., 2020; Gupta and Salgotra, 2022; Ahmed et al., 2023). Additionally, the use of high-throughput sequencing technologies has enabled the identification of epigenetic markers that can be used in breeding programs to select for high-yielding varieties (Yu et al., 2020; Tonosaki et al., 2022).
4.2 The impact of epigenetics on disease resistance
Epigenetic regulation is also pivotal in enhancing disease resistance in wheat. DNA methylation and histone modifications can create "memory marks" that help plants survive various biotic stresses by regulating gene expression based on their epigenetic history (Agarwal et al., 2020; Samantara et al., 2021). For example, CRISPR-based epigenetic editing has been employed to target genes associated with disease resistance, resulting in significantly reduced disease incidence and severity in edited wheat lines (Ahmed et al., 2023). This approach not only improves disease resistance but also contributes to the overall health and productivity of the crop (Gallusci et al., 2017; Duarte-Aké et al., 2023).
4.3 Environmental adaptation and epigenetic regulation
Environmental adaptation in wheat is significantly influenced by epigenetic regulation. Epigenetic modifications enable plants to respond to abiotic stresses such as drought, salinity, and temperature fluctuations by altering gene expression patterns (Agarwal et al., 2020; Samantara et al., 2021). These modifications can be heritable, allowing plants to "remember" past environmental conditions and adapt more efficiently in future generations. The use of epigenetic diversity as a novel source for crop improvement has been highlighted as a potential strategy to develop climate-resilient wheat varieties (Figure 1) (Springer, 2013; Gupta and Salgotra, 2022; Tonosaki et al., 2022). By leveraging epigenetic variations, breeders can enhance the adaptability of wheat to changing environmental conditions, ensuring stable yields and food security (Yu et al., 2020; Duarte-Aké et al., 2023).
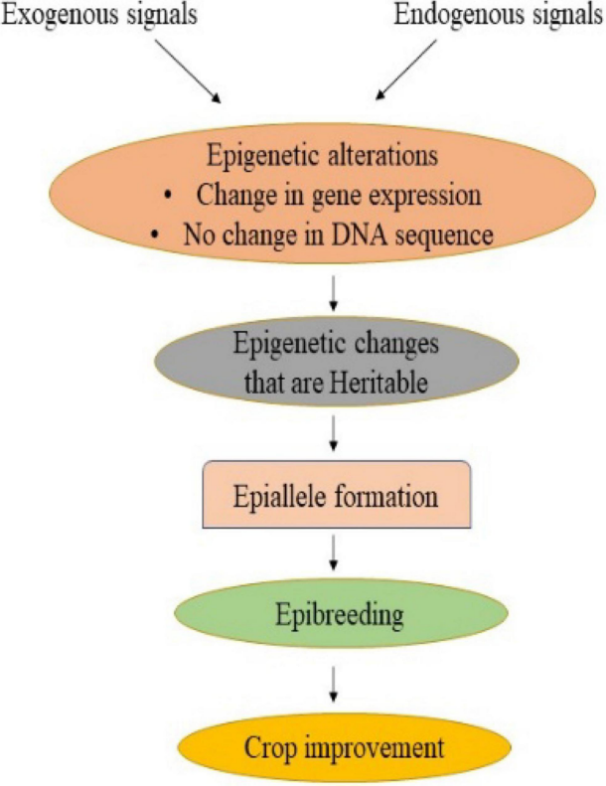 Figure 1 Exogenous and endogenous signals inducing epigenetic variation and their impact on crop breeding in wheat and other crops (Adapted from Gupta and Salgotra, 2022) |
Gupta and Salgotra (2022) found that epigenetics holds great potential in crop breeding. Figure 1 illustrates the pathway of epigenetic variation, beginning with external signals (such as environmental changes) or internal signals (such as gene regulation), which lead to changes in gene expression without altering the DNA sequence. These changes result in heritable epialleles. These epialleles can be used in wheat breeding to target traits such as stress resistance, yield, and quality improvement. As a result, epigenetic breeding represents an emerging strategy for crop improvement, particularly suited to addressing complex environmental challenges such as climate change.
5 The Role of Epigenetics in Wheat Stress Resistance Traits
5.1 Epigenetic regulation of drought resistance traits
Epigenetic mechanisms, such as DNA methylation, play a crucial role in regulating gene expression in response to drought stress in wheat. For instance, the TaBADH-A1 gene, which is involved in osmotic regulation, shows significant expression differences under drought conditions. Wheat cultivars with the BADH-A1b allele exhibit higher expression levels and better drought tolerance compared to those with the BADH-A1a allele, highlighting the importance of epigenetic regulation in drought resistance (Yu et al., 2022). Additionally, plant growth-promoting rhizobacteria (PGPR) have been shown to enhance drought tolerance by modulating the expression of stress-related genes, such as TaDREB2, through epigenetic mechanisms (Barnawal et al., 2017).
A specific case study on the TaBADH-A1 gene demonstrated the potential of DNA methylation in enhancing drought tolerance in wheat. The BADH-A1b allele, associated with higher betaine accumulation, exhibited increased expression under drought stress, leading to improved germination and survival rates. This allele is considered a strong candidate for marker-assisted selection in breeding programs for drought-tolerant wheat varieties. Yu et al. (2022) analyzed the tolerance of different TaBADH-A1 alleles in wheat recombinant inbred lines under drought and salt stress. The results showed that plants containing the BADH-A1b allele (lines 6–10) had significantly higher survival rates under drought and salt stress compared to plants with the BADH-A1a allele (lines 1–5), indicating that the BADH-A1b allele may confer stronger stress resistance (Figure 2). This study provides important evidence for improving drought and salt tolerance in wheat through gene selection, with potential agricultural applications. Additionally, the introduction of the stress-responsive gene SNAC1 from rice into wheat enhanced drought tolerance through epigenetic regulation, further supporting the key role of DNA methylation in stress resistance (Saad et al., 2013).
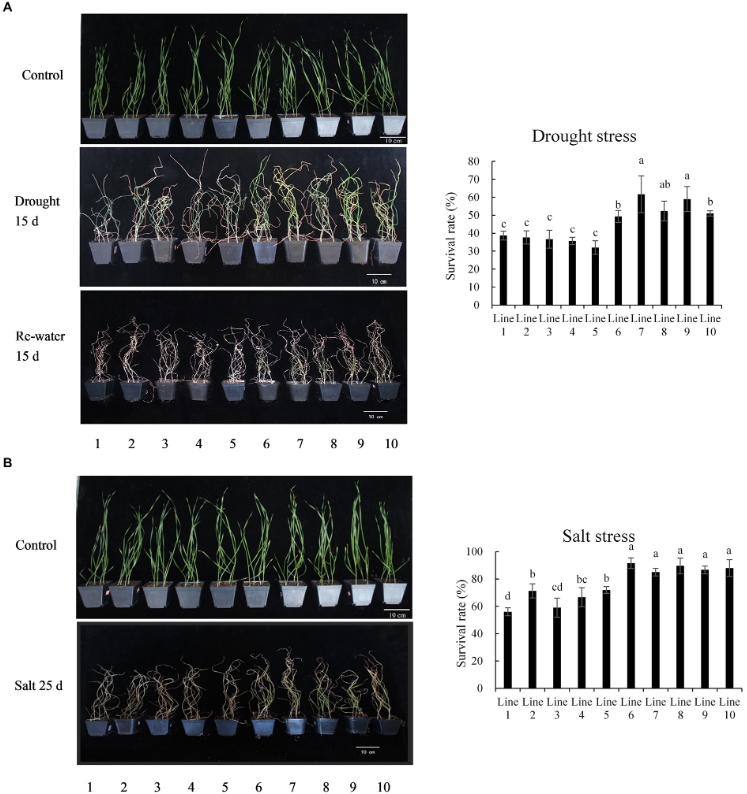 Figure 2 Phenotypic analysis of recombinant inbred lines with different TaBADH-A1 alleles under drought and salt stress (Adapted from Yu et al., 2022) Image caption: A: Survival rate after 5 days of rewatering following 15 days of drought treatment, B: Survival rate after 25 days of irrigation with 700 mM NaCl solution; Lines 1~5 contain the BADH-A1a allele, and lines 6~10 contain the BADH-A1b allele; Different letters indicate statistically significant differences in survival rates (Adapted from Yu et al., 2022) |
5.2 Epigenetic mechanisms of salt and alkaline stress resistance
Salt stress is another major challenge for wheat cultivation, and epigenetic modifications, such as cytosine methylation, have been shown to regulate the expression of key genes involved in salt tolerance. For example, the differential expression of HKT genes, regulated by cytosine methylation, contributes to salt tolerance in wheat. In salt-tolerant genotypes, increased methylation of TaHKT2;1 and TaHKT2;3 genes leads to their downregulation, thereby enhancing salt tolerance (Kumar et al., 2017a; Kumar et al., 2017b). Additionally, the WRKY transcription factors, such as TaWRKY93 and TaWRKY75-A, are regulated epigenetically and play significant roles in mediating salt stress responses by enhancing osmotic adjustment and maintaining membrane stability (Qin et al., 2015; Ye et al., 2021).
5.3 Epigenetics in response to temperature stress
Temperature stress, including both high and low temperatures, affects wheat growth and productivity. Epigenetic mechanisms, such as the regulation of transcription factors, are crucial in mediating temperature stress responses. For instance, the overexpression of TaWRKY93 in Arabidopsis has been shown to enhance tolerance to low temperature stress by upregulating stress-related genes and maintaining higher relative water content and membrane stability (Qin et al., 2015). Similarly, the MYB transcription factor TaMYBsdu1 is differentially regulated under temperature stress, with higher expression levels observed in stress-tolerant genotypes, indicating its potential role in temperature stress adaptation (Rahaie et al., 2010).
6 Potential of Combining Epigenetic Modifications with Wheat Breeding
6.1 Application of epigenetic markers in breeding
Epigenetic markers, such as DNA methylation and histone modifications, have emerged as valuable tools in wheat breeding. These markers can influence gene expression without altering the underlying DNA sequence, providing an additional layer of genetic regulation that can be harnessed for crop improvement. The use of epigenetic markers allows breeders to predict plant performance and increase crop production by taking into account epigenome diversity (Samantara et al., 2021; Gupta and Salgotra, 2022). High-throughput DNA sequencing technologies have enabled the identification of spontaneous epigenetic mutations (epimutations) that contribute to phenotypic diversity, which can be selected for desirable agronomic traits (Tonosaki et al., 2022). This approach complements traditional genetic markers and enhances the efficiency of breeding programs by providing new avenues for selecting traits that improve yield, stress resistance, and overall crop performance (Gallusci et al., 2017; Springer and Schmitz, 2017).
6.2 Utilization of epigenetic variation in non-genetic stable inheritance
Epigenetic variation offers a unique opportunity for crop improvement through non-genetic stable inheritance. Unlike genetic mutations, epigenetic changes can be reversible and do not involve alterations in the DNA sequence. This allows for the creation of new phenotypes that can be inherited over generations, providing a source of variation that can be exploited in breeding programs (Bao, 2008; Tonosaki et al., 2022). The stability and heritability of epigenetic modifications, such as DNA methylation, are crucial for their effective use in breeding strategies. Understanding the mechanisms underlying epigenetic inheritance and their impact on plant development and stress responses can help in developing crops that are better adapted to changing environmental conditions (Gallusci et al., 2017; Latutrie et al., 2019).
Epigenetic variation has been shown to enhance various agronomic traits in wheat, including yield, disease resistance, and stress tolerance. For instance, studies have demonstrated that epigenetic modifications can lead to the development of climate-resilient crops with improved adaptation to changing climates (Samantara et al., 2021; Gupta and Salgotra, 2022). In one case, the application of epigenetic markers in a genome-wide association study (GWAS) identified significant marker-trait associations (MTAs) that explained a substantial portion of phenotypic variance in traits such as yield and disease resistance (Bhatta et al., 2019). These findings highlight the potential of epigenetic variation to create new phenotypes that can be selected for in breeding programs, ultimately leading to the development of elite wheat varieties with enhanced agronomic performance (Dwivedi et al., 2017; Ferreira et al., 2023).
6.3 Integration of gene editing techniques with epigenetics
The integration of gene editing techniques, such as CRISPR/Cas9, with epigenetic modifications holds great promise for advancing wheat breeding. Gene editing allows for precise modifications of the genome, while epigenetic changes can regulate gene expression without altering the DNA sequence. This combination can be used to create targeted epigenetic modifications that enhance desirable traits in wheat (Dwivedi et al., 2017; Tonosaki et al., 2022). For example, epigenome editing can be employed to modify DNA methylation patterns or histone modifications at specific loci, leading to changes in gene expression that improve stress tolerance, yield, and other agronomic traits (Gallusci et al., 2017). By leveraging both genetic and epigenetic tools, breeders can develop more robust and high-performing wheat varieties that meet the demands of a growing population and changing environmental conditions (Bao, 2008; Springer and Schmitz, 2017).
7 Reversibility of Epigenetic Modifications and Their Application in Crop Improvement
7.1 Reversibility of epigenetic modifications
Epigenetic modifications, such as DNA methylation and histone modifications, are crucial for regulating gene expression in plants. These modifications are not permanent and can be reversed, which makes them attractive targets for crop improvement strategies. For instance, histone methylation, a type of post-translational modification, can be dynamically regulated to either maintain or reprogram gene expression, thereby influencing biological outcomes such as development and stress responses (Greer and Shi, 2012). Similarly, DNA methylation levels can be controlled through de novo methylation and active demethylation activities, which are guided by non-coding RNAs and regulated by environmental cues (Kumar and Mohapatra, 2021). The reversibility of these modifications allows for the potential reprogramming of plant traits to enhance crop resilience and productivity.
7.2 Epigenetic plasticity and rapid crop improvement
Epigenetic plasticity refers to the ability of plants to undergo reversible changes in gene expression in response to environmental stimuli without altering the underlying DNA sequence. This plasticity is essential for rapid adaptation to abiotic stresses such as drought, salinity, and extreme temperatures (Liu et al., 2022; Fang et al., 2023). By leveraging epigenetic mechanisms, such as DNA methylation and histone modifications, plants can quickly adjust their physiological and developmental processes to cope with stress conditions. This adaptability can be harnessed to develop stress-resistant crop varieties, thereby facilitating rapid crop improvement. For example, epigenetic modifications have been shown to play a significant role in modulating stress-responsive genes, which can be targeted to enhance crop resilience and yield (Agarwal et al., 2020; Samantara et al., 2021).
7.3 Case study on applying chemical substances to change epigenetic modifications and improve wheat yield
Chemical substances that modify epigenetic marks have been explored as tools to improve crop yield and stress tolerance. For instance, inhibitors of histone deacetylation or DNA methylation have been used in clinical settings to treat diseases, and similar approaches can be applied to agriculture (Kelly et al., 2021). By using these chemical agents, it is possible to induce desirable epigenetic changes that enhance plant growth and yield. In wheat, the application of such chemicals could potentially reprogram the epigenome to improve traits such as drought tolerance, nutrient use efficiency, and overall productivity. This approach offers a promising avenue for crop improvement by directly targeting the epigenetic machinery to achieve rapid and heritable changes in gene expression (Figure 3) (Akhter et al., 2021; Gupta and Salgotra, 2022).
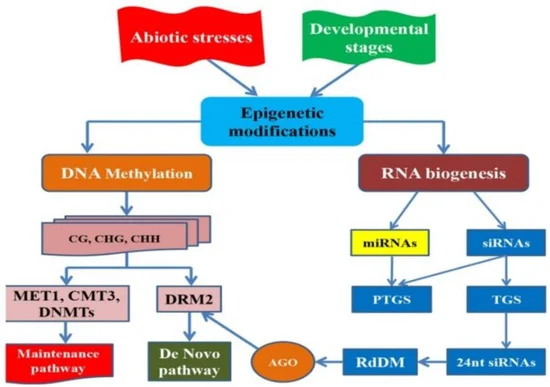 Figure 3 Epigenetic regulatory networks in crop development and responses to abiotic stress (Adapted from Akhter et al., 2021) Image caption: The figure introduces two main layers of epigenetic modifications: DNA methylation and RNA biogenesis; DNA methylation is regulated through maintenance pathways (involving MET1, CMT3, etc.) and de novo methylation pathways (regulated by DRM2); RNA biogenesis controls gene silencing through the production of miRNAs (microRNAs) and siRNAs (small interfering RNAs), which mediate post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS); The RNA-directed DNA methylation (RdDM) pathway further promotes DNA methylation, maintaining gene expression regulation (Adapted from Akhter et al., 2021) |
Akhter et al. (2021) explored how crops such as wheat respond to abiotic stress and control developmental stages through epigenetic modifications. These epigenetic modifications not only regulate gene expression but also ensure crop adaptability to various environmental conditions. DNA methylation and small RNA generation are key pathways in maintaining epigenetic plasticity, enabling rapid responses to abiotic stress through RNA-directed DNA methylation (RdDM) mechanisms. This regulatory mechanism of epigenetics provides powerful tools for crop improvement, particularly in enhancing stress resistance by modulating gene expression in the face of climate change and adverse environmental conditions.
8 Future Development Directions of Wheat Epigenetics
8.1 Application of emerging technologies in wheat epigenetics research
Emerging technologies in the field of epigenetics hold significant promise for advancing wheat crop improvement. High-throughput sequencing technologies, such as whole-genome bisulfite sequencing, have enabled the detailed mapping of DNA methylation patterns across the wheat genome, providing insights into the epigenetic regulation of gene expression (Kapazoglou et al., 2018; Fu, 2024). Additionally, CRISPR/Cas9-based epigenome editing tools are being developed to target specific epigenetic marks, allowing for precise manipulation of gene expression without altering the underlying DNA sequence (Álvarez-Venegas and De-la-Peña, 2016). These technologies can be used to create novel epialleles that confer desirable traits such as increased stress tolerance and improved yield (Saraswat et al., 2017). The integration of these advanced tools into wheat breeding programs could accelerate the development of climate-resilient and high-yielding wheat varieties (Varotto et al., 2020; Kakoulidou et al., 2021).
8.2 Epigenetic regulation and response to environmental changes
Wheat plants are constantly exposed to various environmental stresses, including drought, salinity, and temperature extremes, which can significantly impact their growth and productivity. Epigenetic mechanisms, such as DNA methylation, histone modifications, and non-coding RNAs, play crucial roles in mediating plant responses to these stresses (Kong et al., 2020). For instance, DNA methylation changes have been associated with the regulation of stress-responsive genes, enabling wheat plants to adapt to adverse conditions (Agarwal et al., 2020). Histone modifications, such as acetylation and methylation, also contribute to the dynamic regulation of gene expression in response to environmental cues (Samantara et al., 2021). Understanding these epigenetic responses can provide valuable insights into the development of wheat varieties with enhanced stress tolerance and adaptability to changing climates (Gallusci et al., 2017; Gupta and Salgotra, 2022).
8.3 Synergistic effects of epigenetic modifications and genomic adaptation
The interplay between epigenetic modifications and genomic adaptation is a critical area of research for wheat improvement. Epigenetic changes can influence the expression of genes involved in key agronomic traits, such as flowering time, grain size, and disease resistance, thereby complementing traditional breeding efforts (Álvarez-Venegas and De-la-Peña, 2016). Moreover, the heritability of certain epigenetic marks across generations suggests that epigenetic variation can be harnessed as a stable source of phenotypic diversity (Kapazoglou et al., 2018). By combining genomic selection with epigenetic information, breeders can develop more accurate predictive models for selecting superior wheat genotypes (Gallusci et al., 2017). This synergistic approach has the potential to enhance the efficiency and effectiveness of wheat breeding programs, ultimately leading to the development of high-performing wheat varieties that meet the demands of a growing global population (Saraswat et al., 2017; Varotto et al., 2020; Kakoulidou et al., 2021).
Acknowledgments
Thank you to Ms. Zhou for assisting in organizing a large amount of literature during this research process.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Agarwal G., Kudapa H., Ramalingam A., Choudhary D., Sinha P., Garg V., Singh V., Patil G., Pandey M., Nguyen H., Guo B., Sunkar R., Niederhuth C., and Varshney R., 2020, Epigenetics and epigenomics: underlying mechanisms, relevance, and implications in crop improvement, Functional & Integrative Genomics, 20: 739-761.
https://doi.org/10.1007/s10142-020-00756-7
Ahmed M., Tariq H., Haroon F., Malik S., Dar M., Solangi F., and Aamir S., 2023, Harnessing epigenetic modifications for targeted trait manipulation in pakistani wheat, International Journal of Precision Farming, 1(1): 50-55.
https://doi.org/10.54536/ijpf.v1i1.2227
Akhter Z., Bi Z., Ali K., Sun C., Fiaz S., Haider F., and Bai J., 2021, In response to abiotic stress, DNA methylation confers epigenetic changes in plants, Plants, 10(6): 1096.
https://doi.org/10.3390/plants10061096
Álvarez-Venegas R., and De-la-Peña C., 2016, Editorial: recent advances of epigenetics in crop biotechnology, Frontiers in Plant Science, 7: 413.
https://doi.org/10.3389/fpls.2016.00413
Bao L., 2008, Epigenetic variation and crop genetic improvement, Journal of Jilin Agricultural University, 30(4): 386-393.
Barnawal D., Bharti N., Pandey S., Pandey A., Chanotiya C., and Kalra A., 2017, Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression, Physiologia Plantarum, 161(4): 502-514.
https://doi.org/10.1111/ppl.12614
Bhatta M., Shamanin V., Shepelev S., Baenziger P., Pozherukova V., Pototskaya I., Morgounov A., and Morgounov A., 2019, Marker-trait associations for enhancing agronomic performance, disease resistance, and grain quality in synthetic and bread wheat accessions in western siberia, G3: Genes Genomes Genetics, 9: 4209-4222.
https://doi.org/10.1534/g3.119.400811
Chang Y., Zhu C., Jiang J., Zhang H., Zhu J., and Duan C., 2019, Epigenetic regulation in plant abiotic stress responses, Journal of Integrative Plant Biology, 62(5): 563-580.
https://doi.org/10.1111/jipb.12901
Duarte-Aké F., Us-Camas R., and De-la-Peña C., 2023, Epigenetic regulation in heterosis and environmental stress: the challenge of producing hybrid epigenomes to face climate change, Epigenomes, 7(3): 14.
https://doi.org/10.3390/epigenomes7030014
Dwivedi S., Scheben A., Edwards D., Spillane C., and Ortiz R., 2017, Assessing and exploiting functional diversity in germplasm pools to enhance abiotic stress adaptation and yield in cereals and food legumes, Frontiers in Plant Science, 8: 1461.
https://doi.org/10.3389/fpls.2017.01461
Fang W., Fasano C., and Perrella G., 2023, Unlocking the secret to higher crop yield: the potential for histone modifications, Plants, 12(8): 1712.
https://doi.org/10.3390/plants12081712
Ferreira M., Rocha A., Nascimento F., Oliveira W., Soares J., Rebouças T., Lino L., Haddad F., Ferreira C., Santos-Serejo J., Fernández J., and Amorim E., 2023, The role of somaclonal variation in plant genetic improvement: a systematic review, Agronomy, 13(3): 730.
https://doi.org/10.3390/agronomy13030730
Fu C., 2024, Application of genome-wide association study in crop disease resistance breeding, Field Crop, 7(1): 1-8.
Gallusci P., Dai Z., Génard M., Gauffretau A., Leblanc-Fournier N., Richard-Molard C., Vile D., and Brunel-Muguet S., 2017, Epigenetics for plant improvement: current knowledge and modeling avenues, Trends in Plant Science, 22(7): 610-623.
https://doi.org/10.1016/j.tplants.2017.04.009
Greer E., and Shi Y., 2012, Histone methylation: a dynamic mark in health, disease and inheritance, Nature Reviews Genetics, 13: 343-357.
https://doi.org/10.1038/nrg3173
Gupta C., and Salgotra R., 2022, Epigenetics and its role in effecting agronomical traits, Frontiers in Plant Science, 13: 925688.
https://doi.org/10.3389/fpls.2022.925688
Huang W.Z., 2024, The current situation and future of using GWAS strategies to accelerate the improvement of crop stress resistance traits, Molecular Plant Breeding, 15(2): 52-62.
https://doi.org/10.5376/mpb.2024.15.0007
Jiang Y., N’Diaye A., Koh C., Quilichini T., Shunmugam A., Kirzinger M., Konkin D., Bekkaoui Y., Sari E., Pasha A., Esteban E., Provart N., Higgins J., Rozwadowski K., Sharpe A., Pozniak C., and Kagale S., 2023, The coordinated regulation of early meiotic stages is dominated by non-coding RNAs and stage-specific transcription in wheat, The Plant Journal : for Cell and Molecular Biology, 114(1): 209-224.
https://doi.org/10.1111/tpj.16125
Kakoulidou I., Avramidou E., Baránek M., Brunel-Muguet S., Farrona S., Johannes F., Kaiserli E., Lieberman-Lazarovich M., Martinelli F., Mladenov V., Testillano P., Vassileva V., and Maury S., 2021, Epigenetics for crop improvement in times of global change, Biology, 10(8): 766.
https://doi.org/10.3390/biology10080766
Kapazoglou A., Ganopoulos I., Tani E., and Tsaftaris A., 2018, Epigenetics, epigenomics and crop improvement, Advances in Botanical Research, 86: 287-324.
https://doi.org/10.1016/bs.abr.2017.11.007
Kelly T., Carvalho D., and Jones P., 2010, Epigenetic modifications as therapeutic targets, Nature Biotechnology, 28: 1069-1078.
https://doi.org/10.1038/nbt.1678
Khalid A., Hameed A., and Tahir M., 2023, Wheat quality: a review on chemical composition, nutritional attributes, grain anatomy, types, classification, and function of seed storage proteins in bread making quality, Frontiers in Nutrition, 10: 1053196.
https://doi.org/10.3389/fnut.2023.1053196
Kong L., Liu Y., Wang X., and Chang C., 2020, Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley, International Journal of Molecular Sciences, 21(4): 1480.
https://doi.org/10.3390/ijms21041480
Kumar S., Beena A., Awana M., and Singh A., 2017a, Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes, DNA and Cell Biology, 36: 283-294.
https://doi.org/10.1089/dna.2016.3505
Kumar S., Beena A., Awana M., and Singh A., 2017b, Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance, Frontiers in Plant Science, 8: 1151.
https://doi.org/10.3389/fpls.2017.01151
Kumar S., and Mohapatra T., 2021, Dynamics of DNA methylation and its functions in plant growth and development, Frontiers in Plant Science, 12: 596236.
https://doi.org/10.3389/fpls.2021.596236
Latutrie M., Gourcilleau D., and Pujol B., 2019, Epigenetic variation for agronomic improvement: an opportunity for vegetatively propagated crops, American Journal of Botany, 106: 1281-1284.
https://doi.org/10.1002/ajb2.1357
Li S., Zhang C., Li J., Yan L., Wang N., and Xia L., 2021, Present and future prospects for wheat improvement through genome editing and advanced technologies, Plant Communications, 2 (4): 100211.
https://doi.org/10.1016/j.xplc.2021.100211
Liu Y., Wang J., Liu B., and Xu Z., 2022, Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants, Journal of Integrative Plant Biology, 64(12): 2252-2274.
https://doi.org/10.1111/jipb.13368
Molina-Serrano D., Schiza V., and Kirmizis A., 2013, Cross-talk among epigenetic modifications: lessons from histone arginine methylation, Biochemical Society transactions, 41(3): 751-759.
https://doi.org/10.1042/BST20130003
Pang Y., Liu C., Wang D., Amand P., Bernardo A., Li W., He F., Li L., Wang L., Yuan X., Dong L., Su Y., Zhang H., Zhao M., Liang Y., Jia H., Shen X., Lu Y., Jiang H., Wu Y., Li A., Wang H., Kong L., Bai G., and Liu, S., 2020, High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat, Molecular Plant, 13(9): 1311-1327.
https://doi.org/10.1016/j.molp.2020.07.008
Qin Y., Tian Y., and Liu X., 2015, A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana.. Biochemical and biophysical research communications, 464(2): 428-433.
https://doi.org/10.1016/j.bbrc.2015.06.128
Rahaie M., Xue G., Naghavi M., Alizadeh H., and Schenk P., 2010, A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes, Plant Cell Reports, 29: 835-844.
https://doi.org/10.1007/s00299-010-0868-y
Saad A., Li X., Li H., Huang T., Gao C., Guo M., Cheng W., Zhao G., and Liao Y., 2013, A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses, Plant Science : an International Journal of Experimental Plant Biology, 203-204: 33-40.
https://doi.org/10.1016/j.plantsci.2012.12.016
Samantara, K., Shiv, A., Sousa, L., Sandhu, K., Priyadarshini, P., and Mohapatra, S., 2021, A comprehensive review on epigenetic mechanisms and application of epigenetic modifications for crop improvement, Environmental and Experimental Botany, 188: 104479.
https://doi.org/10.1016/j.envexpbot.2021.104479
Saraswat S., Yadav A., Sirohi P., and Singh N., 2017, Role of epigenetics in crop improvement: water and heat stress, Journal of Plant Biology, 60: 231-240.
https://doi.org/10.1007/s12374-017-0053-8
Saravana Kumar M.S., Wang Y., Zhang X., Cheng H., Sun L., He S., and Hao F., 2020, Redox components: key regulators of epigenetic modifications in plants, International Journal of Molecular Sciences, 21(4): 1419.
https://doi.org/10.3390/ijms21041419
Shafiq S., and Khan A., 2015, Plant epigenetics and crop improvement, In: Barh D., Khan M., Davies E., (eds), PlantOmics: The Omics of Plant Science, Springer, New Delhi, pp.157-179.
https://doi.org/10.1007/978-81-322-2172-2_6
Shewry P., and Hey S., 2015, The contribution of wheat to human diet and health, Food and Energy Security, 4: 178-202.
https://doi.org/10.1002/fes3.64
Shiferaw B., Smale M., Braun H., Duveiller E., Reynolds M., and Muricho G., 2013, Crops that feed the world 10. past successes and future challenges to the role played by wheat in global food security, Food Security, 5: 291-317.
https://doi.org/10.1007/s12571-013-0263-y
Shrawat A., and Armstrong C., 2018, Development and application of genetic engineering for wheat improvement, Critical Reviews in Plant Sciences, 37: 335-421.
https://doi.org/10.1080/07352689.2018.1514718
Springer N., and Schmitz R., 2017, Exploiting induced and natural epigenetic variation for crop improvement, Nature Reviews Genetics, 18: 563-575.
https://doi.org/10.1038/nrg.2017.45
Springer N., 2013, Epigenetics and crop improvement, Trends in Genetics: TIG, 29(4): 241-247.
https://doi.org/10.1016/j.tig.2012.10.009
Tonosaki K., Fujimoto R., Dennis E., Raboy V., and Osabe K., 2022, Will epigenetics be a key player in crop breeding?, Frontiers in Plant Science, 13: 958350.
https://doi.org/10.3389/fpls.2022.958350
Varotto S., Tani E., Abraham E., Krugman T., Kapazoglou A., Melzer R., Radanović A., and Miladinović D., 2020, Epigenetics: possible applications in climate-smart crop breeding, Journal of Experimental Botany, 71: 5223-5236.
https://doi.org/10.1093/jxb/eraa188
Wang M., Li Z., Zhang Y., Zhang Y., Xie Y., Ye L., Zhuang Y., Lin K., Zhao F., Guo J., Teng W., Zhang W., Tong Y., Xue Y., and Zhang Y., 2021, An atlas of wheat epigenetic regulatory elements reveals subgenome divergence in the regulation of development and stress responses, The Plant Cell, 33(4): 865-881.
https://doi.org/10.1093/plcell/koab028
Ye H., Qiao L., Guo H., Guo L., Ren F., Bai J., and Wang Y., 2021, Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-a is referred to drought and salt resistances, Frontiers in Plant Science, 12: 663118.
https://doi.org/10.3389/fpls.2021.663118
Yu J., Xu F., Wei Z., Zhang X., Chen T., and Pu L., 2020, Epigenomic landscape and epigenetic regulation in maize, Theoretical and Applied Genetics, 133: 1467-1489.
https://doi.org/10.1007/s00122-020-03549-5
Yu M., Yu Y., Guo S., Zhang M., Li N., Zhang S., Zhou H., Wei F., Song T., Cheng J., Fan Q., Shi C., Feng W., Wang Y., Xiang J., and Zhang X., 2022, Identification of TaBADH-A1 allele for improving drought resistance and salt tolerance in wheat (Triticum aestivum L.), Frontiers in Plant Science, 13: 942359.
https://doi.org/10.3389/fpls.2022.942359
Zhang Y., Andrews H., Eglitis-Sexton J., Godwin I., Tanurdžić M., and Crisp P., 2022, Epigenome guided crop improvement: current progress and future opportunities, Emerging Topics in Life Sciences, 6: 141-151.
https://doi.org/10.1042/ETLS20210258
.png)
. PDF(566KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jin Zhou
. Xuemei Liu
Related articles
. Wheat
. Epigenetics
. DNA methylation
. Crop improvement
. Gene editing
Tools
. Email to a friend
. Post a comment


