 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2024, Vol. 15, No. 2 doi: 10.5376/tgg.2024.15.0007
Received: 03 Feb., 2024 Accepted: 06 Mar., 2024 Published: 16 Mar., 2024
Xu R.G., and Su Q.S., 2024, Molecular tools and genomic resources in Triticeae: enhancing crop productivity, Triticeae Genomics and Genetics, 15(2): 66-76 (doi: 10.5376/tgg.2024.15.0007)
The Triticeae tribe, which includes essential cereal crops such as wheat and barley, is critical for global food security. Recent advancements in molecular tools and genomic resources have significantly enhanced our ability to improve crop productivity within this tribe. This study explores the latest developments in genomic technologies, genome editing tools, and phenotyping methods that are being utilized to optimize Triticeae crop breeding. Key innovations include haplotype-based approaches for precise genetic diversity identification, open-source genome assembly tools like TRITEX for constructing high-quality genome sequences, and the application of CRISPR/Cas9 for targeted mutagenesis. Additionally, the integration of plant hormonomics for deep physiological phenotyping and high-throughput phenotyping platforms are highlighted as pivotal in understanding and enhancing crop traits. These molecular and genomic advancements collectively contribute to the development of Triticeae crops with improved yield, stress tolerance, and adaptability to changing climates.
1 Introducion
The Triticeae tribe, a significant group within the Poaceae family, encompasses several major cereal crops, including wheat (Triticum spp.), barley (Hordeum vulgare), and rye (Secale cereale) (Chen et al., 2020; Gao et al., 2023). These species are characterized by their large and complex genomes, which have been the focus of extensive genomic research due to their agricultural importance and intricate genetic histories (Rabanus-Wallace and Stein, 2019; Gao et al., 2023). Advances in sequencing technologies have facilitated the assembly of high-quality reference genomes and pan-genome studies, providing deeper insights into the genetic diversity and evolutionary dynamics of Triticeae species (Rabanus-Wallace and Stein, 2019; Gao et al., 2023).
Triticeae crops are vital for global food security, serving as staple foods for a significant portion of the world's population and as essential sources of animal feed and industrial raw materials (Sakuma and Schnurbusch, 2020; Khalid et al., 2023). Wheat alone accounts for approximately half of the food calories consumed worldwide, highlighting its critical role in human nutrition (Khalid et al., 2023). The genetic and genomic advancements in Triticeae have enabled the identification of key traits related to yield, stress tolerance, and disease resistance, which are crucial for improving crop productivity and sustainability (Hensel, 2019; Sakuma and Schnurbusch, 2020; Kuluev et al., 2022). The application of modern biotechnological tools, such as CRISPR/Cas genome editing, has further accelerated the development of improved Triticeae varieties with enhanced agronomic traits (Hensel, 2019; Kuluev et al., 2022).
This study provides a comprehensive overview of molecular tools and genomic resources for Triticeae crops, focusing on enhancing crop productivity. It summarizes advancements in genome sequencing and pan-genome analyses, discusses the genetic and molecular bases of key agronomic traits like yield potential and stress tolerance, and highlights the role of genome editing technologies in breeding programs. Future research directions and the integration of genomic resources for sustainable agriculture and food security are also explored. By synthesizing recent findings, the study offers insights into genetic and genomic strategies to enhance Triticeae crop productivity and resilience, contributing to global agricultural sustainability.
2 Molecular Tools in Triticeae
2.1 Genomic sequencing technologies
Recent advancements in next-generation sequencing (NGS) technologies have significantly reduced the cost of DNA sequencing, enabling the sequencing of species with large and complex genomes, such as bread wheat (Triticum aestivum L.) (Hussain et al., 2022). The international effort to sequence the wheat genome has culminated in the release of a fully annotated reference wheat-genome assembly, which has paved the way for the pan-genomic era in wheat research. This has facilitated the use of genotyping arrays capable of characterizing hundreds of wheat lines with thousands of markers, providing fast, relatively inexpensive, and reliable data for wheat breeding (Hussain et al., 2022). Additionally, open-source computational workflows like TRITEX have been developed to construct chromosome-scale sequence assemblies, further enhancing genomic studies in Triticeae crops (Monat et al., 2019).
2.2 Marker-assisted selection (MAS)
Marker-assisted selection (MAS) has revolutionized plant breeding by improving the productivity and accuracy of classical breeding methods. DNA markers have been extensively used to map quantitative trait loci (QTLs) and associate them with specific genes, thereby accelerating the development of new crop varieties (Hasan et al., 2021). Functional markers (FMs) closely associated with phenotypic traits have been identified using various genomics approaches, facilitating the direct selection of genes associated with desirable traits (Salgotra and Stewart, 2020). MAS has been successfully applied in breeding programs to improve traits such as drought resilience in alfalfa, demonstrating its potential to enhance crop productivity under stress conditions (Singh et al., 2022).
2.3 CRISPR-Cas9 and genome editing
CRISPR-Cas9 has emerged as a powerful, cost-effective, and versatile tool for precise and efficient genome editing in plants. Unlike earlier genome editing tools like zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR-Cas9 involves simpler designing and cloning methods, making it more accessible for targeted mutagenesis (Jaganathan et al., 2018; Razzaq et al., 2019). This technology has been used for single base substitution, multiplex gene editing, gene knockouts, and regulation of gene transcription, significantly contributing to crop improvement (Razzaq et al., 2019). The development of CRISPR-Cas12a and other advanced CRISPR systems has further enhanced the efficiency and specificity of genome editing, enabling the engineering of multiple genes simultaneously (Liu et al., 2019). These advancements promise to bridge the gap between forward and reverse genetics, providing rapid methods to validate genes and alleles for crop improvement (Thomson et al., 2022).
2.4 Transcriptomics and gene expression profiling
Functional genomics approaches, including transcriptomics, have been instrumental in identifying genes and functional markers associated with plant phenotypic variation. Transcriptomics involves the comprehensive analysis of gene expression profiles, which helps in understanding the molecular mechanisms underlying various traits (Salgotra and Stewart, 2020). This approach, combined with other functional genomics strategies like TILLING and association mapping, has been used to identify candidate genes linked to key traits such as grain yield, end-use quality, and resistance to biotic and abiotic stresses in wheat (Hussain et al., 2022). The integration of transcriptomics with other molecular tools provides a holistic view of gene function and regulation, facilitating the development of improved crop varieties.
In summary, the integration of genomic sequencing technologies, marker-assisted selection, CRISPR-Cas9 genome editing, and transcriptomics has significantly advanced the molecular breeding of Triticeae crops. These molecular tools offer new opportunities to enhance crop productivity and resilience, addressing the challenges of global food security.
3 Genomic Resources in Triticeae
3.1 Reference genomes and genome assemblies
The development of high-quality reference genomes and genome assemblies has been pivotal in advancing Triticeae research. The TRITEX workflow, an open-source computational tool, has significantly improved the assembly of large-genome Triticeae crops such as wheat and barley. This tool integrates various sequencing data types to construct sequence scaffolds with high contiguity, which are then ordered into chromosomal pseudomolecules. TRITEX has been successfully applied to tetraploid wild emmer, hexaploid bread wheat, and the barley cultivar Morex, providing valuable resources for the research community (Monat et al., 2019). Additionally, the generation of multiple wheat genome assemblies has revealed extensive structural variations and gene content differences among wheat lines, which are crucial for understanding the genetic basis of traits and for breeding purposes (Walkowiak et al., 2020). The release of a gold-standard, fully annotated reference wheat-genome assembly in 2018 marked a significant milestone, enabling more efficient exploitation of genomic resources in wheat breeding (Hussain et al., 2022).
3.2 Genomic databases and bioinformatics tools
The establishment of genomic databases and bioinformatics tools has facilitated the analysis and interpretation of complex Triticeae genomes. GeneTribe, a novel homology inference method, incorporates gene collinearity to connect emerging genome assemblies within the Triticeae tribe. This tool has been integrated into the Triticeae-GeneTribe database, which includes 12 Triticeae genomes and 3 outgroup model genomes, providing versatile analysis and visualization functions (Chen et al., 2022) (Figure 1). Furthermore, the Zymoseptoria tritici ORFeome library, which includes over 3000 quality-controlled plasmids, represents a powerful resource for functional genomics studies, offering opportunities to understand the biology of this wheat pathogen (Chaudhari et al., 2019).
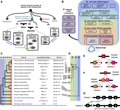 Figure 1 Challenges and strategies for homolog inference in the Triticeae tribe in the pangenomic era (Adopted from Chen et al., 2020) Image caption: (A) Frequent polyploidization events shaped the complex relationships between Triticeae species; (B) Multiple-level scenarios for pairwise comparison of assemblies as centered by IWGSC RefSeqv1.1. Triticum–Aegilops is considered a combined genus; (C) Hierarchical organization of currently published high-quality genome assemblies that are available in the TGT database. (D) Polypoid genomes are decomposed into multiple diploid subgenomes for pairwise comparison; (E) A model showing that each assembly of variety 1 and variety 2 has one well-annotated gene (filled box); these genes would be computationally considered orthologs derived from one gene in an ancient species. The unfilled box indicates a duplicated pseudogene that is not annotated or has lost its function; (F) A modified model in which the assembly of the middle species is available and an ancient gene duplication event followed by reciprocal loss of function was uncovered. Gene A1 and gene a2 originated from the paralogous genes Am and gene am in the middle species, respectively; (G) A model showing that gene loss events or incomplete gene annotations hinder accurate orthology inference between closely related genomes. (Adopted from Chen et al., 2020) |
Chen et al. (2020) addresses the challenges of homology inference in the Triticeae tribe, shaped by frequent polyploidization and reticulate evolution. It introduces a multi-level framework to organize assemblies and annotations, enabling efficient gene-gene mapping and hierarchical database construction. The framework classifies pairwise relationships into six levels, applying different strategies for each to enhance accuracy. The study highlights the limitations of sequence similarity-based methods, particularly for genetically similar assemblies, and incorporates additional information such as collinearity, annotation quality, and chromosome groups to improve inference accuracy. By decomposing polyploid genomes into diploid subgenomes and considering gene loss and duplication events, the proposed method aims to provide a more reliable inference of homology relationships. This approach enhances our understanding of Triticeae genomics and supports the development of more accurate genomic tools for crop improvement.
3.3 Genetic diversity and germplasm collections
Efficient utilization of genetic diversity in germplasm collections is essential for crop improvement. Bioinformatic approaches have been developed to extract functional genetic diversity from heterogeneous germplasm collections. For instance, genome-wide SNP data can capture significantly more haplotypic diversity compared to traditional methods based on geographic or environmental data. This approach has been demonstrated in crops like sorghum, where subsets of germplasm collections were assembled to maximize diversity at functional loci relevant to breeding programs (Reeves et al., 2020). Additionally, a haplotype-led approach has been developed to increase the precision of wheat breeding by identifying genetic diversity using genome assemblies from multiple wheat cultivars. This method allows for the focused discovery of novel haplotypes, enhancing the efficiency and precision of breeding efforts (Brinton et al., 2020).
3.4 Functional genomics resources
Functional genomics resources are crucial for understanding gene function and improving crop traits. The high-resolution genome-wide association study (GWAS) in wheat has identified numerous quantitative trait loci (QTL) and candidate genes associated with important agronomic traits. This study utilized a large panel of wheat cultivars and high-density SNP markers, providing a solid foundation for QTL fine mapping and candidate gene validation (Pang et al., 2020). Additionally, the development of ORFeome libraries, such as the one for Zymoseptoria tritici, enables the functional characterization of genes and their encoded proteins, facilitating the study of gene function and interactions (Chaudhari et al., 2019). Advances in genome engineering technologies, such as CRISPR/Cas, further enhance the potential for functional genomics studies and the development of improved Triticeae varieties (Hensel, 2019).
By leveraging these genomic resources and tools, researchers can enhance the productivity and resilience of Triticeae crops, addressing the challenges posed by a growing global population and changing environmental conditions.
4 Applications of Molecular Tools in Enhancing Crop Productivity
4.1 Development of disease-resistant varieties
The development of disease-resistant crop varieties is crucial for ensuring food security and sustainable agriculture. Molecular tools, particularly CRISPR/Cas genome editing, have revolutionized the breeding of disease-resistant crops. CRISPR/Cas systems have been effectively used to modify genes responsible for resistance to various biotic stresses, including fungal, viral, and bacterial pathogens. For instance, recent advances in CRISPR/Cas-mediated genome editing have enabled the rapid development of crop varieties with enhanced resistance to multiple biotic stresses, significantly reducing crop losses and improving yield stability (Hensel, 2019; Wang et al., 2022; Parihar et al., 2022). The integration of quantitative trait loci (QTLs) and marker-assisted selection further accelerates the breeding process by allowing precise introgression of resistance genes from different sources (Parihar et al., 2022).
4.2 Enhancing abiotic stress tolerance
Abiotic stresses such as drought, salinity, extreme temperatures, and heavy metal toxicity pose significant challenges to crop productivity. Molecular tools, especially CRISPR/Cas9, have been instrumental in developing crop varieties with enhanced tolerance to these stresses. CRISPR/Cas9 allows for precise editing of genes involved in abiotic stress responses, thereby improving the resilience of crops under adverse environmental conditions (Jaganathan et al., 2018; Surabhi et al., 2019; Nascimento et al., 2023; Joshi et al., 2023). The use of ribonucleoproteins (RNPs) as a DNA-free strategy for genome editing has also emerged as a promising approach to avoid regulatory hurdles associated with genetically modified organisms (GMOs) (Nascimento et al., 2023). Additionally, the integration of functional genomics and transcriptomics has expanded our understanding of the molecular mechanisms underlying abiotic stress tolerance, facilitating the development of superior-performing genotypes (Joshi et al., 2023).
4.3 Improving yield and quality traits
Improving yield and quality traits is a primary objective in crop breeding programs. Molecular tools such as CRISPR/Cas9 have enabled the precise modification of genes associated with yield and quality traits, leading to the development of high-yielding and nutritionally superior crop varieties. For example, CRISPR/Cas9 has been used to enhance traits such as grain size, nutrient content, and stress tolerance, contributing to increased yield potential and improved crop quality (Jaganathan et al., 2018; Wang et al., 2022; Medina-Lozano and Díaz, 2022). The exploration of crop wild relatives (CWRs) as a source of genetic diversity has also been pivotal in introducing desirable traits into cultivated varieties, thereby broadening the genetic base and enhancing crop performance (Mammadov et al., 2018).
4.4 Accelerating breeding programs
The integration of molecular tools in breeding programs has significantly accelerated the development of improved crop varieties. Techniques such as CRISPR/Cas9 genome editing, marker-assisted selection, and genomic selection have streamlined the breeding process by enabling precise and efficient modification of target genes (Hensel, 2019; Wang et al., 2022; Parihar et al., 2022). The availability of comprehensive genetic maps and reliable DNA markers has facilitated the rapid introgression of desirable traits, reducing the time required to develop new varieties (Parihar et al., 2022). Additionally, advancements in genome editing technologies have opened new avenues for creating super cultivars with broad resistance to biotic and abiotic stresses, thereby enhancing crop productivity and sustainability (Jaganathan et al., 2018; Wang et al., 2022).
In summary, the application of molecular tools such as CRISPR/Cas9 has revolutionized crop breeding by enabling the development of disease-resistant varieties, enhancing abiotic stress tolerance, improving yield and quality traits, and accelerating breeding programs. These advancements hold great promise for meeting the growing global food demand and ensuring agricultural sustainability.
5 Case Studies
5.1 Successful examples in wheat improvement
Recent advancements in genomic tools have significantly contributed to the improvement of wheat (Triticum aestivum L.). One notable example is the development of TRITEX, an open-source computational workflow that has enhanced the assembly of chromosome-scale genome sequences in wheat. This tool has been instrumental in constructing high-quality reference genome sequences, which are crucial for pan-genomic studies and have facilitated the identification of genomic regions associated with important agronomic traits (Monat et al., 2019) (Figure 2).
 Figure 2 TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools (Adopted from Monat et al., 2019) Image caption: Morex V1 pseudomolecules, an improved BAC-by-BAC assembly, and a genome-wide optical map; Panel (a) shows a dot plot alignment revealing high collinearity at a megabase scale but discordance at finer resolutions; Panel (b) presents a more consistent alignment with the improved Dovetail BAC-by-BAC assembly, showing a higher correlation and concordance in orientations; Panel (c) aligns optical map contigs to TRITEX scaffolds, indicating high-confidence matches but highlighting some unaligned restriction sites (Adapted from Monat et al., 2019) |
Monat et al. (2019) validates the TRITEX assembly method by comparing it with various genomic resources, demonstrating its high accuracy and reliability. The TRITEX super-scaffolds showed strong concordance with the Morex V1 pseudomolecules, improved BAC-by-BAC assemblies, and the genome-wide optical map, indicating robust sequence alignment and improved assembly quality. This validation highlights TRITEX's effectiveness in producing precise and comprehensive genome sequences, addressing previous assembly issues, and enhancing our understanding of the Triticeae genomes. The findings emphasize TRITEX's potential to significantly advance genomic studies and breeding programs in Triticeae crops, contributing to more accurate and efficient genetic research and crop improvement strategies.
Additionally, a high-resolution genome-wide association study (GWAS) identified 395 quantitative trait loci (QTL) for 12 traits in wheat, using a panel of 768 cultivars. This study not only pinpointed known genes and QTL but also identified eight putative candidate genes that enhance spike seed-setting andgrain size. These findings are pivotal for genomics-based breeding programs aimed at improving wheat yield (Pang et al., 2020).
Moreover, the integration of wild genetic resources into wheat breeding programs has been explored to introduce new diversity for genes or alleles of agronomical interest. This approach has the potential to accelerate the development of new improved cultivars, although it is often hampered by the genetic background of the recipient varieties or the donor (Laugerotte et al., 2022).
5.2 Advances in barley genomics
Barley (Hordeum vulgare) has also seen significant advancements in genomic research. The TRITEX workflow has been applied to barley, resulting in an improved annotated reference genome sequence assembly of the barley cultivar Morex. This assembly serves as a valuable community resource for further genomic studies (Monat et al., 2019).
Furthermore, the use of the CRISPR/Cas genome editing system has been a breakthrough in barley research. This system has enabled precise genetic modifications, which are essential for studying gene functions and improving agronomic traits. The majority of studies have focused on bread wheat and barley, highlighting the potential of CRISPR/Cas technology in these crops (Kuluev et al., 2022).
5.3 Emerging research in rye and Triticale
Rye (Secale cereale) and Triticale (× Triticosecale Wittm.) have also benefited from recent genomic advancements. The genetic control of compatibility in crosses between wheat and its wild or cultivated relatives, including rye, has been extensively studied. This research has led to the identification of genetic factors controlling crossing ability, which is crucial for the creation of triticale. The application of modern genomics technologies in breeding programs is expected to accelerate the improvement of both wheat and triticale (Laugerotte et al., 2022).
In addition, the development of the Triticeae-GeneTribe database, which integrates 12 Triticeae genomes and 3 outgroup model genomes, has provided a valuable resource for homology inference and analysis. This database has facilitated the study of complex evolutionary histories and structural rearrangements in Triticeae crops, including rye and triticale (Chen et al., 2020).
Overall, these case studies highlight the significant progress made in the genomic research of Triticeae crops, which is essential for enhancing crop productivity and meeting the demands of a growing global population.
6 Challenges and Future Directions
6.1 Technical and scientific challenges
The advancement of genomic tools and molecular technologies in Triticeae, particularly wheat, faces several technical and scientific challenges. The hexaploid nature of wheat (Triticum aestivum) complicates genetic research due to its large and complex genome, which includes significant gene redundancy (Li et al., 2021). This complexity hinders precision gene modifications and the breeding of elite cultivars. Additionally, the integration of multi-omics data, such as genomics, transcriptomics, and metabolomics, remains a significant challenge due to the vast amount of data and the need for sophisticated computational tools to analyze and interpret these datasets (Pathak et al., 2018; Chen et al., 2021). The development of open-source tools like TRITEX for chromosome-scale sequence assembly is a step forward, but further improvements are needed to enhance the accuracy and efficiency of these assemblies (Monat et al., 2019).
6.2 Integrating multi-omics data
Integrating multi-omics data is crucial for a comprehensive understanding of the molecular mechanisms underlying important agronomic traits in Triticeae. Multi-omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, have been successfully applied to various crops, providing insights into growth, yield, and stress responses (Yang et al., 2021). However, the integration of these datasets poses significant challenges due to the complexity and volume of the data. Advanced computational methods and systems biology approaches are required to construct predictive models that can link genotype to phenotype and vice versa (Chen et al., 2021; Yang et al., 2021). For instance, the integration of sRNAome, transcriptome, and degradome data in T. turgidum has provided valuable insights into the regulatory networks of grain development and stress response, highlighting the potential of multi-omics approaches in crop improvement (Liu et al., 2020).
6.3 Future prospects in Triticeae genomics
The future of Triticeae genomics lies in the continued development and application of advanced genomic and molecular tools. Genome editing technologies, such as CRISPR/Cas, offer promising prospects for precise genetic modifications and the development of improved crop varieties (Kuluev et al., 2022). The use of haplotype-based approaches and high-resolution genome-wide association studies (GWAS) can enhance the precision of breeding programs by identifying and exploiting genetic diversity (Brinton et al., 2020; Pang et al., 2020). Additionally, the integration of multi-omics data with systems biology approaches can facilitate the discovery of functional genes and the development of robust models for predicting complex traits (Chen et al., 2021; Yang et al., 2021). These advancements will be crucial for addressing the challenges of global food security and environmental sustainability.
6.4 Policy and regulatory considerations
The successful implementation of advanced genomic tools and molecular technologies in Triticeae requires supportive policy and regulatory frameworks. Policies should promote the adoption of innovative technologies while ensuring the safety and sustainability of genetically modified crops. Regulatory considerations should address the ethical, environmental, and socio-economic impacts of genome editing and other molecular breeding techniques (Li et al., 2021; Kuluev et al., 2022). International collaboration and harmonization of regulatory standards will be essential to facilitate the global exchange of knowledge and resources, thereby accelerating the development and deployment of improved Triticeae cultivars. Additionally, policies should support research and development initiatives, capacity building, and the dissemination of knowledge to ensure that the benefits of these technologies reach all stakeholders, including smallholder farmers and developing countries (Pathak et al., 2018).
7 Concluding Remarks
The research on molecular tools and genomic resources in Triticeae has demonstrated significant advancements in enhancing crop productivity. The development and application of molecular marker-assisted selection (MAS) have revolutionized plant breeding by increasing the accuracy and speed of developing new crop varieties. High-throughput genotyping platforms and next-generation sequencing (NGS) technologies have enabled the sequencing of complex genomes, such as that of bread wheat, facilitating the identification of quantitative trait loci (QTLs) and candidate genes associated with key agronomic traits. The integration of genomic selection (GS) and CRISPR/Cas9-mediated gene editing has further accelerated the breeding process, allowing for the precise manipulation of genetic variation to improve yield, stress resistance, and quality traits.
Future research should focus on the continued development and refinement of genomic tools and technologies to further enhance the efficiency and effectiveness of crop improvement programs. The integration of omics approaches, such as transcriptomics and proteomics, with traditional breeding methods will provide a more comprehensive understanding of the genetic basis of important traits. Additionally, the exploration of pan-genomic resources and the development of associative transcriptomics platforms will enable the identification of novel genetic markers and candidate genes for trait improvement. Research should also prioritize the development of climate-resilient crop varieties through the application of GS and advanced gene-editing techniques, addressing the challenges posed by climate change and ensuring global food security.
The advancements in molecular tools and genomic resources have significantly contributed to the progress in Triticeae crop improvement. The integration of MAS, GS, and CRISPR/Cas9 technologies has opened new avenues for precise and efficient breeding, ultimately enhancing crop productivity and resilience. Continued investment in genomic research and the development of innovative breeding strategies will be crucial in meeting the growing demands for food in the face of global challenges. The collaborative efforts of researchers, breeders, and policymakers will be essential in translating these scientific advancements into practical solutions for sustainable agriculture.
Acknowledgments
The authors extend our sincere thanks to two anonymous peer reviewers for their invaluable feedback on the initial draft of this paper.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Brinton J., Ramírez-González R., Simmonds J., Wingen L., Orford S., Griffiths S., Haberer G., Spannagl M., Walkowiak S., Pozniak C., and Uauy C., 2020, A haplotype-led approach to increase the precision of wheat breeding, Communications Biology, 3: 712.
https://doi.org/10.1038/s42003-020-01413-2
Chen J., Xue M., Liu H., Fernie A., and Chen W., 2021. Exploring the genic resources underlying metabolites through mGWAS and mQTL in wheat: From large-scale gene identification and pathway elucidation to crop improvement, Plant Communications, 2(4): 100216.
https://doi.org/10.1016/j.xplc.2021.100216
PMid:34327326 PMCid:PMC8299079
Chen Y., Song W., Xie X., Wang Z., Guan P., Peng H., Jiao Y., Ni Z., Sun Q., and Guo W., 2020, A Collinearity-incorporating homology inference strategy for connecting emerging assemblies in Triticeae tribe as a pilot practice in the plant pangenomic era, Molecular Plant, 13(12): 1694-1708.
https://doi.org/10.1016/j.molp.2020.09.019
PMid:32979565
Chaudhari Y., Cairns T., Sidhu Y., Attah V., Thomas G., Csukai M., Talbot N., Studholme D., and Haynes K., 2019. The Zymoseptoria tritici ORFeome: a functional genomics community resource, Molecular Plant-Microbe Interactions: MPMI, 32: 12.
https://doi.org/10.1094/MPMI-05-19-0123-A
PMid:31272284
Gao Z., Bian J., Lu F., Jiao Y., and He H., 2023, Triticeae crop genome biology: an endless frontier, Frontiers in Plant Science, 14: 1222681.
https://doi.org/10.3389/fpls.2023.1222681
PMid:37546276 PMCid:PMC10399237
Hasan N., Choudhary S., Naaz N., Sharma N., and Laskar R., 2021. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes, Journal of Genetic Engineering & Biotechnology, 19(1): 128.
https://doi.org/10.1186/s43141-021-00231-1
PMid:34448979 PMCid:PMC8397809
Hensel G., 2019. Genetic transformation of Triticeae cereals-Summary of almost three-decade's development, Biotechnology Advances, 40: 107484.
https://doi.org/10.1016/j.biotechadv.2019.107484
PMid:31751606
Hussain B., Akpınar B., Alaux M., Algharib A., Sehgal D., Ali Z., Aradottir G., Batley J., Bellec A., Bentley A., Cagirici H., Cattivelli L., Choulet F., Cockram J., Desiderio F., Devaux, P., Doğramacı, M., Dorado, G., Dreisigacker, S., Edwards, D., El-Hassouni, K., Eversole, K., Fahima, T., Figueroa, M., Gálvez S., Gill K., Govta L., Gul A., Hensel G., Hernández P., Crespo-Herrera L., Ibrahim A., Kilian B., Korzun V., Krugman T., Li,Y., Liu S., Mahmoud A., Morgounov A., Muslu T., Naseer F., Ordon F., Paux E., Perović D., Reddy G., Reif J., Reynolds M., Roychowdhury R., Rudd J., Sen T., Sukumaran S., Ozdemir B., Tiwari V., Ullah N., Unver T., Yazar S., Appels R., and Budak H., 2022, Capturing wheat phenotypes at the genome level, Frontiers in Plant Science, 13: 851079.
https://doi.org/10.3389/fpls.2022.851079
PMid:35860541 PMCid:PMC9289626
Jaganathan D., Ramasamy K., Sellamuthu G., Jayabalan S., and Venkataraman G., 2018, CRISPR for crop improvement: an update review, Frontiers in Plant Science, 9: 985.
https://doi.org/10.3389/fpls.2018.00985
PMid:30065734 PMCid:PMC6056666
Joshi A., Yang S., Song H., Min J., and Lee J., 2023, Genetic databases and gene editing tools for enhancing crop resistance against abiotic stress, Biology, 12(11): 1400.
https://doi.org/10.3390/biology12111400
PMid:37997999 PMCid:PMC10669554
Khalid A., Hameed A., and Tahir M., 2023. Wheat quality: A review on chemical composition, nutritional attributes, grain anatomy, types, classification, and function of seed storage proteins in bread making quality, Frontiers in Nutrition, 10: 1053196.
https://doi.org/10.3389/fnut.2023.1053196
PMid:36908903 PMCid:PMC9998918
Kuluev B., Mikhailova E., Kuluev A., Galimova A., Zaikina E., and Khlestkina E., 2022, Genome editing in species of the tribe Triticeae with the CRISPR/Cas System, Molekuliarnaia Biologiia, 56(6): 949-968.
https://doi.org/10.1134/S0026893322060127
Laugerotte J., Baumann U., and Sourdille P., 2022, Genetic control of compatibility in crosses between wheat and its wild or cultivated relatives, Plant Biotechnology Journal, 20: 812-832.
https://doi.org/10.1111/pbi.13784
PMid:35114064 PMCid:PMC9055826
Li S., Zhang C., Li J., Yan L., Wang N., and Xia, L., 2021, Present and future prospects for wheat improvement through genome editing and advanced technologies, Plant Communications, 2(4): 100211.
https://doi.org/10.1016/j.xplc.2021.100211
PMid:34327324 PMCid:PMC8299080
Liu H., Able A., and Able J., 2020, Multi-omics analysis of small RNA, transcriptome, and degradome in T. turgidum-regulatory networks of grain development and abiotic stress response, International Journal of Molecular Sciences, 21(20): 7772.
https://doi.org/10.3390/ijms21207772
PMid:33096606 PMCid:PMC7589925
Liu Q., Zhang Y., Li F., Li J., Sun W., and Tian C., 2019, Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila, Biotechnology for Biofuels, 12: 293.
https://doi.org/10.1186/s13068-019-1637-y
PMid:31890021 PMCid:PMC6927189
Mammadov J., Buyyarapu R., Guttikonda S., Parliament K., Abdurakhmonov I., and Kumpatla S., 2018, Wild relatives of maize, rice, cotton, and soybean: treasure troves for tolerance to biotic and abiotic stresses, Frontiers in Plant Science, 9: 886.
https://doi.org/10.3389/fpls.2018.00886
PMid:30002665 PMCid:PMC6032925
Medina-Lozano I., and Díaz A., 2022, Applications of genomic tools in plant breeding: crop biofortification, International Journal of Molecular Sciences, 23(6): 3086
https://doi.org/10.3390/ijms23063086
PMid:35328507 PMCid:PMC8950180
Monat C., Padmarasu S., Lux T., Wicker T., Gundlach H., Himmelbach A., Ens J., Li C., Muehlbauer G., Schulman A., Waugh R., Braumann I., Pozniak C., Scholz U., Mayer K., Spannagl M., Stein N., and Mascher M., 2019, TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools, Genome Biology, 20: 284.
https://doi.org/10.1186/s13059-019-1899-5
PMid:31849336 PMCid:PMC6918601
Nascimento F., Rocha A., Soares J., Mascarenhas M., Ferreira M., Lino L., Ramos A., Diniz L., Mendes T., Ferreira C., Santos-Serejo J., and Amorim E., 2023, Gene editing for plant resistance to abiotic factors: a systematic review, Plants, 12(2): 305.
https://doi.org/10.3390/plants12020305
PMid:36679018 PMCid:PMC9860801
Pang Y., Liu C., Wang D., Amand P., Bernardo A., Li W., He F., Li L., Wang L., Yuan X., Dong L., Su Y., Zhang H., Zhao M., Liang Y., Jia H., Shen X., Lu Y., Jiang H., Wu Y., Li A., Wang H., Kong L., Bai G., and Liu S., 2020, High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat, Molecular Plant, 13(9): 1311-1327.
https://doi.org/10.1016/j.molp.2020.07.008
PMid:32702458
Parihar A., Kumar J., Gupta D., Lamichaney A., Sj S., Singh A., Dixit G., Gupta S., and Toklu F., 2022, Genomics enabled breeding strategies for major biotic stresses in pea (Pisum sativum L.), Frontiers in Plant Science, 13: 861191.
ttps://doi.org/10.3389/fpls.2022.861191
PMid:35665148 PMCid:PMC9158573
Pathak R., Baunthiyal M., Pandey D., and Kumar A., 2018, Augmentation of crop productivity through interventions of omics technologies in India: challenges and opportunities, 3 Biotech, 8: 1-28.
https://doi.org/10.1007/s13205-018-1473-y
PMid:30370195 PMCid:PMC6195494
Razzaq A., Saleem F., Kanwal M., Mustafa G., Yousaf S., Arshad H., Hameed M., Khan M., and Joyia F., 2019, Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 Toolbox, International Journal of Molecular Sciences, 20(16): 4045.
https://doi.org/10.3390/ijms20164045
PMid:31430902 PMCid:PMC6720679
Rabanus-Wallace M., and Stein N., 2019, Progress in sequencing of Triticeae genomes and future uses, Applications of Genetic and Genomic Research in Cereals, England, pp.19-47.
https://doi.org/10.1016/B978-0-08-102163-7.00002-8
Reeves P., Tetreault H., and Richards C., 2020. Bioinformatic extraction of functional genetic diversity from heterogeneous germplasm collections for crop improvement, Agronomy, 10(4): 593.
https://doi.org/10.3390/agronomy10040593
Salgotra R., and Stewart C., 2020, Functional markers for precision plant breeding, International Journal of Molecular Sciences, 21(13): 4792.
https://doi.org/10.3390/ijms21134792
PMid:32640763 PMCid:PMC7370099
Sakuma S., and Schnurbusch T., 2020, Of floral fortune: tinkering with the grain yield potential of cereal crops, The New Phytologist, 225(5): 1873-1882.
https://doi.org/10.1111/nph.16189
PMid:31509613
Singh L., Pierce C., Santantonio N., Steiner R., Miller D., Reich J., and Ray I., 2022, Validation of DNA marker‐assisted selection for forage biomass productivity under deficit irrigation in alfalfa, The Plant Genome, 15(1): e20195.
https://doi.org/10.1002/tpg2.20195
PMid:35178866
Surabhi G., Badajena B., and Sahoo S., 2019, Genome editing and abiotic stress tolerance in crop plants, In: Wani S., (eds) Recent approaches in omics for plant resilience to climate change, Springer, Cham, pp.35-56.
https://doi.org/10.1007/978-3-030-21687-0_2
Thomson M., Biswas S., Tsakirpaloglou N., and Septiningsih E., 2022, Functional allele validation by gene editing to leverage the wealth of genetic resources for crop improvement, International Journal of Molecular Sciences, 23(12): 6565.
https://doi.org/10.3390/ijms23126565
PMid:35743007 PMCid:PMC9223900
Walkowiak S., Gao L., Monat C., Haberer G., Kassa M., Brinton J., Ramírez-González R., Kolodziej M., Delorean E., Thambugala D., Klymiuk V., Byrns B., Gundlach H., Bandi V., Siri J., Nilsen K., Aquino C., Himmelbach A., Copetti D., Ban T., Venturini L., Bevan M., Clavijo B., Koo D., Ens J., Wiebe K., N’Diaye A., Fritz A., Gutwin C., Fiebig A., Fosker C., Fu B., Accinelli G., Gardner K., Fradgley N., Gutierrez-Gonzalez J., Halstead-Nussloch G., Hatakeyama M., Koh C., Deek J., Costamagna A., Fobert P., Heavens D., Kanamori H., Kawaura K., Kobayashi F., Krasileva K., Kuo T., McKenzie N., Murata K., Nabeka Y., Paape T., Padmarasu S., Percival‐Alwyn L., Kagale S., Scholz U., Sese J., Juliana P., Singh R., Shimizu‐Inatsugi R., Swarbreck D., Cockram J., Budak H., Tameshige T., Tanaka T., Tsuji H., Wright J., Wu J., Steuernagel B., Small I., Cloutier S., Keeble-Gagnère G., Muehlbauer G., Tibbets J., Nasuda S., Melonek J., Hucl P., Sharpe A., Clark M., Legg E., Bharti A., Langridge P., Hall A., Uauy C., Mascher M., Krattinger S., Handa H., Shimizu K., Distelfeld A., Chalmers K., Keller B., Mayer K., Poland J., Stein N., McCartney C., Spannagl M., Wicker T., and Pozniak C., 2020, Multiple wheat genomes reveal global variation in modern breeding, Nature, 588: 277-283.
https://doi.org/10.1038/s41586-020-2961-x
PMid:33239791 PMCid:PMC7759465
Wang Y., Zafar N., Ali Q., Manghwar H., Wang G., Yu L., Ding X., Ding F., Hong N., Wang G., and Jin S., 2022, CRISPR/Cas genome editing technologies for plant improvement against biotic and abiotic stresses: advances, limitations, and future perspectives, Cells, 11(23): 3928.
https://doi.org/10.3390/cells11233928
PMid:36497186 PMCid:PMC9736268
Yang Y., Saand M., Huang L., Abdelaal W., Zhang J., Wu Y., Li J., Sirohi M., and Wang F., 2021, Applications of multi-omics technologies for crop improvement, Frontiers in Plant Science, 12: 563953.
https://doi.org/10.3389/fpls.2021.563953
PMid:34539683 PMCid:PMC8446515
.png)
. PDF(553KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Rugang Xu
. Qiuxia Su
Related articles
. Triticeae
. Genomic resources
. CRISPR/Cas9
. Phenotyping
. Crop productivity
Tools
. Email to a friend
. Post a comment


