Analysis of Characteristics, Mutant Sites and Evolution of Dehydrin 6 (DHN6) Protein in Three Types of Rowed Barleys 
2. Department of Cell Biology and Genetics, Zunyi Medical College, Zunyi, 563099, P.R. China
3. Department of Obstetrics and Gynecology, Affiliated Hospital, Zunyi Medical College, Zunyi, 563099, P.R. China
 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2012, Vol. 3, No. 1 doi: 10.5376/tgg.2012.03.0001
Received: 10 Feb., 2012 Accepted: 12 Mar., 2012 Published: 13 Mar., 2012
Liu et al., 2012, Analysis of Characteristics, Mutant Sites and Evolution of Dehydrin 6 (DHN6) Protein in Three Types of Rowed Barleys, Triticeae Genomics and Genetics, Vol.3, No.1 1-8 (doi: 10.5376/tgg.2012.03.0001)
Dehydrins (DHNs), a special polypeptide generated in late embryogenesis of higher plants, could protect the plants from the damage caused by cell dehydration. In order to learn the relationship between characteristics and functions of dehydrins, we cloned Dhn6 genes from three types of rowed barleys, and bioinformatics analysis showed that they encoded proteins composed of 523 (six-rowed barley), 502 (four-rowed barley) and 486 (two-rowed barley) amino acid residues, respectively. Furthermore, analysis of amino acid mutations found that there were whole conservative traits and mutant sites specificity in this gene. Analyses of protein characteristics and the secondary structure indicated that DHN6 was a highly hydrophilic alkaline protein, and linear structure and numerous random curls were the main component of secondary structure. Moreover, K-segment was involved in the formation of the α-helix, which presumed that the amphipathic α-helices domain of DHN6 might play important roles in protecting membrane structure during the hydration process. The construction of phylogenetic tree of 21 species in this study showed that Dhn6 gene could be an efficient foundation for identifying and distinguishing of different species associated with special sequences of nucleotides, and had a closer genetic distance in Gramineae crops.
Water deficit, the most limiting factor of plant growth and crop production, induces various biochemical and physiological responses in plants (Kiani et al., 2007). Plants respond to water deficit through multiple physiological mechanisms at the cellular, tissue, and whole-plant levels. These responses are not only dependent on the severity and duration of the water deficit, but also on the developmental stage and morphological/anatomical parameter of the plants (Ludlow and Muchow, 1990; Smith and Griffiths, 1993). Late embryogenesis abundant (LEA) proteins were believed to play a significant role in the stress response in various organisms including plants, algae, yeasts and bacteria (Ramanjulu and Bartels, 2002). Dehydins (DHNs), molecular weight 9~20 kD, are among the most frequently observed proteins in plants under water stress (Suprunova et al., 2004). In the barley genome, recent investigations into dehydrin multigene family have been identified 13 Dhn genes, which encode 4 sub classes of DHNs: YnSKn, SKn, Kn and KS, respectively, based on permutations in the arrangement of characterized domains (Campbell and Close, 1997; Werner-Fraczek and Close, 1998; Rodriguez et al., 2005).
In a few studies, Garay-Arroyo et al (2000) and Qian et al (2010) argued the review that amphipathic α-helices formed by K-sequence could play an important role in protecting membrane structure. Association between tolerance to stresses involving dehydration (drought, salinity or freezing) and accumulation of members of the Dhn family has been established in different species such as wheat, barley poplar and sunflower (Giordani et al., 1999; Lopez et al., 2003). Genetic variability in the stress response has been suggested to be mainly due to the differential expression of stress-responsive genes (Joshi et al., 1997). Many genes respond experimentally to water stress, however, their precise functions either in tolerance or sensitivity often remain unclear (Ludlow and Muchow, 1990; Smith and Griffiths, 1993). It is thus critical to study functions of stress-induced genes to understand the mechanisms involved in stress tolerance in plants. Correlating phenotypic adaptations with molecular characters should enable us to evaluate the role of dehydrins during adaptation (Guo et al., 2009). Therefore, the possible relationship between diverse molecular trait of DHN6 and phylogenesis was investigated in two-, four- and six-rowed barley lines in this research.
1 Results and Analysis
1.1 Cloning and sequence analyses of Dhn6
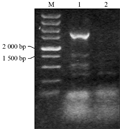
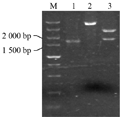
Taken the barley genomic DNA of BZ-26, BZ-16, BZ-12, respectively as the template, we amplified the target genes. The results of agarose gel electrophoresis validation showed that the specific fragments between 1 500 bp and 2 000 bp in length can be obtained, when the annealing temperature was 55℃ (Figure 1). Identification of the recombinant of BZ-12 was further digested with EcoRâ… and Hindâ…¢ (Figure 2). Sequence analyses found that the length in BZ-26, BZ-16 and BZ-12 was 1 657 bp, 1 705 bp and 1 767 bp, respectively, and the sequence alignment displayed that the homology among them reached 93.08%. Moreover, the Dhn6 sequence in two-rowed barley (BZ-26) was the shortest, apart from absence 42 bp nucleotide sequences at +583 bp , as well as at +787 bp lacking TGGTGC and at +908 bp lacking GGTGTC starting from ATG. In addition, another 47 nucleotide sites appeared mutation. Compared with BZ-26, BZ-16 was surplus 48 bp nucleotides at +583 bp and +787 bp, additionally, there were 11 base sites appeared mutation. However, the surplus of 63 bp appeared at +791 bp in six-rowed barley (BZ-12). Using the online software Blastn to retrieve the GenBank database, the results revealed that the Dhn6 gene sequence in this paper could not be exactly matched with any sequence that has been submitted, though the sequence homology was very high, which indicated that Dhn6 could be applied to identify species.
|
|
|
|
1.2 Characteristics of DHN6 amino acid sequences
Analyses of open reading frame (ORF) with ORF finder showed the ORF in BZ-26 was shortest about 1 458 bp in length, encoding 486 amino acid residues, and that of BZ-16 and BZ-12 was about 1 506 bp, 502 amino acid residues and 1 569 bp and 523 amino acid residues, respectively. The amino acid sequence analyses displayed that the deduced protein (Y2SK2 type) was composed of the highly conversed motifs including in Y-sequence (2), S-sequence (1) and K-sequence (2) in three typical rowed barleys (Figure 3).
Comparing amino acid sequences of DHN6 of different rowed barley with that of hull-less barley (GenBank accession No. AF043091), we found 21 mutant sites in the deduced protein in those three types in the present study. From figure 3, we found that the encoded dehydrin protein sequence in the six-rowed barley, BZ-12, was the longest, containing a peptide enriched Gly (GYGGGVTGTGTTGTHGTGHTT), whereas, that of BZ-26 was the shortest, and lacking 18 amino acid residues. Among these three types of rowed barleys, there were 63 mutation sites totally, the lowest mutant ratio 7.94% (5/63) was detected in the third base of genetic code, and that of 17.46% (11/63) in the second genetic code position. However, the highest proportion of mutants 19.05% (12/63) was taken place in the first base of genetic code. Therefore, it was concluded that the mutation frequency of three sites bias was differential (Table 1).
|
|
|
|
1.3 Characteristics and secondary structures of DHN6
Analyses of physical and chemical characteristics of three types of rowed barley found that DHN6, a stable and highly hydrophilic basic protein, was composed of 18 amino acid residues, and enriched Gly, but was absent of Trp and Cys (Table 2). The results showed that the molecular weight of DHN6 in BZ-12 was the largest, in despite of containing a Gly peptide, but the Gly content and hydrophilic index is not the highest, whereas, the hydropathicity parameter in BZ-26 and BZ-16 was the highest, and BZ-26 was the stablest. Therefore, hydropathicity index was not related to instability index, molecular weight and contents of Gly.
|
|
To predict the relationship between characteristic and secondary structure, we analyzed the secondary structure of DHN6 among two-, four-, and six-rowed barley (Table 3). The results showed that the secondary structure of DHN6 involving four basic structures such as alpha helix, beta turn, extrended strand, and random coil. We also knew that from table 3 the percentage of extrended strand and random coil was highest, reached up to 80%, they were the main component of DHN6 secondary structure. Furthermore, The highest ratio of alpha helix and random coil was observed in two-rowed barley (BZ-26), was 7.41% and 60.29%, respectively. Moreover, three alpha helixes formed by K-sequence appeared in the different rowed barley and hull-less barley (Figure 4).
|
|
|
|
1.4 Phylogenetic analysis of amino acid sequences in DHN6
Based on the characteristic sequences and conserved motifs of DHN6 in GenBank, we selected 21 species to construct the phylogenetic tree and study the relationships of phylogenetic and molecular evolutionary, selecting the highest scores of E-values in the same species. As shown in Figure 5, three scaffolds of those species were consisted in the molecular evolutionary, and the tested materials were attributed to the same branch including GU216698 and hull-less barley (AF043091). Notably, the separated genetic relationship was detected in the woody plants such as Prunus persica (CAC00637), Pinus sylvestris (CAD54622) and Picea abies (ABU89751).
|
|
2 Discussion
In general, the conserved amino acid mutations were usually determined by the conserved nucleotides. Namely, the slower evolutionary speed was frequently observed in the higher bias base of the species (Tamura, 1992; Sun et al., 2008). In this study, analysis of amino acid mutations of DHN6 indicated that sequence similarity was consistent to the conserved nucleotide sites in barley (Table 1). Moreover, we found that higher conserved nucleotide sites appeared in the first site of genetic sites of Dhn6 gene. In fact, due to the environmental choice pressure, changeable replacement sites were determined by bias base of genetic codes. The characteristics of conserved sites and bias base of genetic codes showed that higher ratio of replacement/transversion could take place in Dhn6 gene. It was accordingly deduced that DHNs would be played an important role in evolutionary process of plants suffered from water deficit.
DHNs might act as water attractants in cells with low water potential, having a role in osmotic potential regulation based on the characters of amino acid sequences (Compbell and Close, 1997; Porcel et al., 2005). In our tests, none of contents of Gly, molecular weight, instability index (Table 2), and secondary structure (Table 3) was associated with hydropathicity index in the deduced protein of DHN6. It was a possible conclusion that hydropathicity capacity of DHNs was attributed to the advanced structures of protein. Reports indicated DHNs could counteract the irreversible damaging effects of increasing ionic strength in the cytosol during desiccation by sequestration of ions in plants under water stress (Close, 1997; Danyluk et al., 1998). In recent experiment, Qian et al (2011) argued a view that amphipathic α-helices of DHN6 might perform a physical and stable protection of peripheral membrane in plant cells subjected to water deficit and temperature changes. Here, the authors described that amphipathic α-helices formed by the conserved motifs of N-, C-, and K-sequence could be associated with peripheral membrane protections in the water-deficit cell.
Generally, mtRNA and rRNA of the eukaryotic cells were applied to analyze the phylogenetic characters in the most of studies (Jia et al., 2007; Emre et al., 2007; Marschner et al., 2007). However, inaccurate conclusions could be obtained from the experimental materials because of their highly variable eco-geographic origins. Sun et al (2008), supporting our views, also agreed with an argument that functional genes could be served as phylogenetic relationships in the sampled eco-geographic plants.
In our tests, the same scaffold of barley was accurately observed in the gramineae crops, as compared with the far genetic distance in the woody plants such as Prunus persica, Pinus sylvestris and Picea abies (Figure 5). As described above, earlier genetic separation took place in barleys while the other scaffolds performed their respective evolutions involving Triticum turgidum ssp. Durum, Zea mays and Oryza sativa. The results of phylogenetic relationships indicated that Dhn6 gene could be an efficient foundation for identifying and distinguishing of different species associated with those special amino acid sequences (Figure 5). Moreover, phylogenetic tree may elucidate not only the molecular evolutionary relationships among diverse species, but the functional roles based on amino acid sequences of the deduced proteins (Khuri et al., 2001; Xiong et al., 2011). The last step in the dehydration-signaling cascade was the alternation of genes responsible for the synthesis of compounds that serve to protect cellular structures against the deleterious effects of dehydration, such as proteins with protective functions encoded for by the late embryogenesis abundant (LEA) genes (Bartels and Souer, 2004; Hazen et al., 2005). Therefore, it was concluded that Dhn6 gene could be used as a reference for identification of species, associated with the important protective roles and phylogenetic tree of DHN6.
3 Materials and Methods
3.1 Plant materials
The experimental materials, harvested from Yun-Gui plateau, were selected to compare with their molecular traits, secondary structure and phylogeny of DHN6, including six-rowed barley (BZ-12), four-rowed line (BZ-16) and two-rowed one (BZ-26) in the present study.
3.2 Extraction of genomic DNA and cloning of Dhn6 gene
Total genomic DNA was extracted from 7-day-old fresh seedlings following a modified cetyltrimethyl ammonium bromide (CTAB) protocol, as described by Saghai-Maroof et al (1984). PCR reactions were performed in a volume of 20 µL containing 50 ng genomic DNA, 1 × PCR buffer, 200 µM dNTPs, 10 mM of primers (P1: 5'-CGGCATCCGCTTGACATT-3', and P2: 5'-GCAAGTCAGGCTCAGTTCAGT-3') and 0.5 U of Taq polymerase (TOYOBO Co., LTD., China). PCR reaction was started at denaturation step of 10 min, followed by 35 cycles at 95℃ for 60 s, 55℃ for 60 s, 72℃ for 60 s, and terminated at 72℃ for 8 min. PCR products were separated on 1.2% agarose gels and purified with DNA gel extraction Kit. The purified PCR products were cloned into plasmid vector pMD18-T (TaKaRa Biotechnology Co., Ltd.) and followed sequenced for each genotype in triplicate.
3.3 Prediction and analysis of protein structural domain
Sequence similarity analysis in GenBank was performed using the Blast 2.1 search tool (http://www.ncbi.nlm.nih.gov/blast/). Nucleotides and amino acid sequence analyses were performed with DNAMAN programme. To predict the biophysics characteristics of the putative protein of DHN6, software on the ExPASy Proteomics Server (http://au.expasy.org/) was used. The prediction and analysis for the protein structural domain and functional site were performed using Prosite software (http://www.expasy.org/prosite/), involving molecular weight, theoretical isoelectric point (pI) and character of amino acid sequences. Phylogenetic and molecular evolutionary analyses were conducted using MEGA 3.1 software (Kumar et al., 2004). ClustalW software (http://www.ebi.ac.uk/clustalw/) was used for alignment of multiple sequences.
Authors' Contributions
MSL performed the experiment and wrote the article; QZ and JJP performed the experiment of gene cloning and data analysis; ZZ analyzed the data of other parts; GQ was responsible for the project and modified the manuscript. All authors have read and approved the final manuscript.
Acknowledgements
We express our deep appreciations to the financial supports of this research from Guizhou Province Natural Sciences Foundation of China (J-2007-2050), the Key Project of Zunyi City Natural Sciences Foundation of China (N-2007-05).
References
Bartels D., and Souer E., 2004, Molecular responses of higher plants to dehydration, In Plant Responses to Abiotic Stress (eds., H. Hirt; K. Shinozaki), Springer-Verlag, Berlin, Heidelberg, pp.9-38
Campbell, S.A., and Close, T.J., 1997, Dehydrins: genes, proteins, and associated with phenotypic traits, New Phytologist, 137(1): 61-74
http://dx.doi.org/10.1046/j.1469-8137.1997.00831.x
Close T.J., 1997, Dehydrins: a commonality in the response of plants to dehydration and low temperatures, Physiologia Plantarum, 100(2): 291-296
http://dx.doi.org/10.1034/j.1399-3054.1997.1000210.x
http://dx.doi.org/10.1111/j.1399-3054.1997.tb04785.x
Danyluk J., Perron A., Houde M., Limin A., Fowler B., Benhamou N., and Sarhan F., 1998, Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat, Plant Cell, 10(4): 623-638
http://dx.doi.org/10.2307/3870737 PMid:9548987 PMCid:144014
http://dx.doi.org/10.1105/tpc.10.4.623
Emre Y., Hurtaud C., Ricquier D., Bouillaud F., Hughes J., and Criscuolo F., 2007, Avian UCP: the killjoy in the evolution of the mitochondrial uncoupling proteins, Journal of Molecular Evolution, 65(4): 392-402
http://dx.doi.org/10.1007/s00239-007-9020-1 PMid:17909695
Garay-Arroyo A., Colmenero-Flores J.M., Garciarrubio A., and Covarrubias A.A., 2000, Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit, Journal of Biological Chemistry, 275(8): 5668-5674
http://dx.doi.org/10.1074/jbc.275.8.5668 PMid:10681550
Giordani, T., Natali, L., Ercole, A.D., Pugliesi, C., Fambrini, M., Vernieri, P., Vitagliano, C., and Cavallini, A., 1999, Expression of a dehydrin gene during embryo development and drought stress in ABA-deficient mutants of sunflower (Helianthus annuus L.), Plant Molecular Biology, 39(4): 739-748
http://dx.doi.org/10.1023/A:1006194720022 PMid:10350088
Guo P., Baum M., Grando S., Ceccarelli S., Bai G., Li R., Korff M.V., Varshney R.K., Graner A., and Valkoun J., 2009, Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage, Journal of Experimental Botany, 60(12): 3531-3544
http://dx.doi.org/10.1093/jxb/erp194 PMid:19561048 PMCid:2724701
Hazen S.P., Pathan M.S., Sanchez A., Baxter I., Dunn M., Estes B., Chang H.S., Zhu T., Kreps J.A., and Nguyen H.T., 2005, Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array, Functional Integrative Genomics, 5(2): 104-116
http://dx.doi.org/10.1007/s10142-004-0126-x
Jia S., Chen H., Zhang G., Wang Z., Lei C., Yao R., and Han X., 2007, Genetic variation of mitochondrial d-loop region and evolution analysis in some Chinese cattle breeds. Journal of Genetics and Genomics, 34(6): 510-518
http://dx.doi.org/10.1016/S1673-8527(07)60056-3
Joshi, C.P., Kluveva, N.Y., Morrow, K.J., and Nguyen, H.T., 1997, Expression of a unique plastid localized heat shock protein is genetically linked to acquired thermotolerance in wheat, Theoretical and Applied Genetics, 95(5-6): 834-841
http://dx.doi.org/10.1007/s001220050633
Khuri S., Bakker F.T., and Dunwell J.M., 2001, Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins, Molecular Biology and Evolution, 18(4): 593-605
http://dx.doi.org/10.1093/oxfordjournals.molbev.a003840 PMid:11264412
Kiani, S.P., Grieu, P., Maury, P., Hewezi, T., Gentzbittel, L., and Sarrafi, A., 2007, Genetic variability for physiological traits under drought conditions and differential expression of water stress-associated genes in sunflower (Helianthus annuus L.), Theoretical and Applied Genetics, 114(2): 193-207
http://dx.doi.org/10.1007/s00122-006-0419-7 PMid:17103138
Kumar S., Tamura K., and Nei M., 2004, MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment, Brief Bioinform, 5(2): 150-163
http://dx.doi.org/10.1093/bib/5.2.150 PMid:15260895
Lopez, C.G., Banowetz, G.M., Peterson, C.J., and Kronstad, W.E., 2003, Dehydrin expression and drought tolerance in seven wheat cultivars, Crop Science, 43(2): 577-582
http://dx.doi.org/10.2135/cropsci2003.0577
Ludlow, M.M., and Muchow, R.C., 1990, A critical evaluation of traits for improving crop yields in water-limited environments, Advances in Agronomy, 43: 107-153
http://dx.doi.org/10.1016/S0065-2113(08)60477-0
Marschner S., Meister A., Blattner F.R., and Houben A., 2007, Evolution and function of B chromosome 45S rDNA sequences in Brachycome dichromosomatica, Genome, 50(7): 638-644
http://dx.doi.org/10.1139/G07-048 PMid:17893741
Porcel R., Azcón R., and Ruiz-Lozano J.M., 2005, Evaluation of the role of genes encoding for dehydrin proteins (LEA D-11) during drought stress in arbuscular mycorrhizal Glycine max and Lactuca sativa plants, Journal of Experimental Botany, 56(417): 1933-1942
http://dx.doi.org/10.1093/jxb/eri188 PMid:15911559
Qian G., Ping J.J., Zhang Z., Luo S.Y., Li X.Y., Yang M.Z., and Zhang D., 2011, Molecular cloning and protein structure prediction of barley (Hordeum vulgare L.), Dhn6 gene and its expression pattern under dehydration conditions, Hereditas, 33(3): 270-277 PMid:21402536
Ramanjulu S., and Bartels D., 2002, Drought- and desiccation-induced modulation of gene expression in plants, Plant, Cell and Environment, 25(2): 141-151
http://dx.doi.org/10.1046/j.0016-8025.2001.00764.x PMid:11841659
Rodriguez, E.M., Svensson, J.T., Malatrasi, M., Choi, D.W., and Close, T.J., 2005, Barley Dhn13 encodes a KS-type dehydrin with constitutive and stress responsive expression, Theoretical and Applied Genetics, 110(5): 852-858
http://dx.doi.org/10.1007/s00122-004-1877-4 PMid:15711789
Saghai-Maroof M.A.S., Soliman K.M., Jorgensen R.A., and Allard R.W., 1984, Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics, Proceedings of the National Academy of Sciences of the USA, 81(24): 8014-8018
http://dx.doi.org/10.1073/pnas.81.24.8014
Smith, J.A.C., and Griffiths, H., 1993, Water deficits: plant responses from cell to community, Bios Scientific Publishers, Oxford, UK, pp.1-332
Sun D.Q., Jiang Y.J., Han X.Y., Qu Y.Y., Bi Y.H., and Zhang G.H., 2008, Cloning and phylogenetic analysis of bovine prochymosin gene, Hereditas, 30(7): 863-869
PMid:18779129
Suprunova, T., Krugman, T., Fahima, T., Chen, G., Shams, I., Korol, A., and Nevo, E., 2004, Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit, Plant Cell Environ, 27(10): 1297-1308
http://dx.doi.org/10.1111/j.1365-3040.2004.01237.x
Tamura K., Nei M., and Kumar S., 2004, Prospects for inferring very large phylogenies by using the neighbor-joining method, Proceedings of the National Academy of Sciences of the USA, 101(30): 11030-11035
http://dx.doi.org/10.1073/pnas.0404206101 PMid:15258291 PMCid:491989
Tamura K., 1992, The rate and pattern of nucleotide substitution in drosophila mitochondrial DNA, Molecular Biology and Evolution, 9(5): 814-825
PMid:1528108
Werner-Fraczek, J.E., and Close, T.J., 1998, Genetic studies of Triticeae dehydrins: assignment of seed proteins and a regulatory factor to map positions, Theoretical and Applied Genetics, 97(1-2): 220-226
http://dx.doi.org/10.1007/s001220050888
Xiong G.J., Xu Q., and Hua J.P., 2011, Isolation and expression analysis of two GhBlind homologs in upland cotton (Gossypium hirsutum L.), Acta Agronomica Sinica, 37(2): 362-368
http://dx.doi.org/10.3724/SP.J.1006.2011.00362
. PDF(612KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Maosheng Liu
. Qian Zhou
. Junjiao Ping
. Zhen Zhang
. Gang Qian
Related articles
. Barley ( Hordeum vulgare L.)
. Dhn 6 gene
. Sequence alignment
. Secondary structure
. Phylogenesis
Tools
. Email to a friend
. Post a comment

.png)

.png)


