2 Northeast Agricultural University, Harbin, 150030, P.R. China
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2011, Vol. 2, No. 3 doi: 10.5376/lgg.2011.02.0003
Received: 13 Oct., 2011 Accepted: 12 Dec., 2011 Published: 16 Dec., 2011
Tang et al., 2009,Heat Shock Factor 8 Introducing into Soybean (Glycine max) by Agrobacterium-Mediated Transformation, Molecular Plant Breeding, 7(3): 444-450 (doi: 10.3969/mpb.007.000444)
Environmental stress seriously affects growth and development of crops. Heat shock transcription factor (heat shock factor 8, HSF8) is a class of proteins that play important roles in the reactions of heat shock, the main functions of heat shock gene are binding the corresponding heat shock element in the process of expression, initiating the gene transcription process and, ultimately, facilitating the expression of heat shock protein (HSP) gene. In this paper, we inserted hsf8 gene into dicotyledon expression vector pCAMBIA3300 that contains the selection marker, bar gene. The construct, named pCAMBIA3300-HSF8, has been transfered into new soybean lines, Hajiao5337 and Hajiao5489, mediated by Agrobacterium tumefaciens. In the practice of soybean genetic transformation, we explored the impact factors on the transformation of soybean cotyledon mediated by Agrobacterium, and optimized the transformation conditions. It is the way to increase the selection efficiency by delaying the screening after co-culture with optimal concentration of glufosinate-ammonium 3.5 mg/L in selection culture medium. We obtained the T1 generation transgenic plants with pCAMBIA3300-HSF8 by Agrobacterium-mediated aprroch derived from Hajiao5337 and Hajiao5489 , of which 17 plants were proved by PCR to be positive transgenic plants. Furthermore, transcription level of hsf8 gene in T1 transgenic plants with glufosinate-ammonium resistance was detected by Real-time PCR approach, the results showed that 9 of 17 plants had much higher expression level than that of the reference of Hajiao5337, whereas one transgenic plant had lower level than that of another reference Hajiao5489.
Soybean is the world's most important grain and oil crops. Total annual production of soybean in China could not meet the increasing domestic consumer’s demand, which would contribute to the low yield of soybean. Increasing the level of soybean yield depends on the soybean varieties with the resistant abilities to adverse growing conditions and with the effective use of nutrients. Environmental stresses such as drought, soil salinity and extreme temperatures and so on are serious impact on crop growth and development, resulting in yield decrease (Wang et al., 2003), of which the adverse temperature is dominating factor. In fact, Soybean is a kind of crop lack of high temperature tolerance, the temperature is too high or too dry at the flowering and pod developing stage that will lead to soybean yield reducing. It was found that synthesis of heat shock proteins (heat stress / shock protein, HSP) was positively correlated to acquiring biological heat tolerance. HSP synthesis can improve the response ability of organisms, particularly in the capacity of heat tolerance (Zhu et al., 2006).It is said that many organisms receiving heat shock treatment under the below of the lethal temperature could improve the survival opportunity in the face of adversity.
Expression and regulation of Heat shock protein gene may be achieved by heat shock transcription factor (HSF), which mainly occurs at the transcriptional level. In higher organisms, potential HSF might be a potential activator in the heat shock environment, which would be a process along with the intra- and inter-molecular interactions of HSF as well as the interaction between HSF and heat shock / HSE (heat stress shock elements). In addition, HSFs as a kind of stress signal response transcription factors, also showed some resistance to cold and drought. There are some evidences that plant HSFs have broadly cross-protections under the different stress conditions (Lee et al., 1995; Lee and Schöffl, 1996; Busch et al., 2005).
In this study, we ligated the hsf8 gene into pCAMBIA3300, a dicotyledonous plant expression vector, by using soybean cotyledonary-node as explant, the hsf8 gene was introduced into two novel lines, Hajiao5337 and Hajiao5489, by Agrobacterium-mediated transformation. Real-time PCR method was employed to determine the T1 transgenic plants for obtaining the hsf8 gene highly expressing in transgenic plants. The objectives of this study were to achieve and improve expression levels of some soybean target genes such as HSP70 through expressing heat shock transcription factor hsf8, to enhance soybean tolerance to high temperature stress, and to provide novel germplasm to develop stress-resistant soybean varieties.
1 Results and Analysis
1.1 Construction of hsf8 gene expression vector
Plant gene expression vector was constructed following technical scheme shown in Figure 1. Plasmid pBI121-HSF8 was digested with EcoRâ… and Hind â…¢ to be recovered in the size of approximate 2.8 kb fragment (Figure 2, lane 1), which contained the completely 35S + hsf8 + NOS expression structure. Plasmid pCAMBIA3300 also digested with EcoR â… and Hind â…¢ restriction enzymes, about 8.5 kb fragment was recovered (Figure 2, lane 2), and then ligated both recovered fragments. The ligated products were transformed into E. coli DH5α. The recombinants were screened with 100 mg/L Kanamycin. The candidate recombinants were validated by digestion of EcoR â… and Hind â…¢ restriction enzyme (Figure 2, lane 3), which can regenerate the same size of pBI121-HSF8 and pCAMBIA3300, named pCAMBIA3300-HSF8.
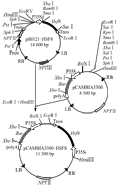 Figure 1 The scheme for constructing transformation vector containing hsf8 gene |
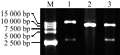 Figure 2 Plant expression vector pCAMBIA3300-HSF8 validated with endonucleases Note: M: DL15000; 1: pBI121-HSF8 digested with EcoRâ… and Hindâ…¢; 2: pCAMBIA3300 digested with EcoRâ… and Hindâ…¢; 3: pCAMBIA3300-HSF8 digested with EcoRâ… and Hindâ…¢ |
1.2 Selective pressure of glufosinate-ammonium on soybean explants
According to preliminary results, concentrations of glufosinate-ammonium were set up as 0 mg/L, 0.5 mg/L, 1 mg/L, 1.5 mg/L, 2 mg/L, 2.5 mg/L, 3 mg/L, 3.5 mg/L and 4 mg/L. in order to study the effects of glufosinate-ammonium on the budding rate of explants (Figure 3). The results showed the differential rate of cotyledonary-node of Hajiao5337 and Hajiao5489 was 6.7% that was significantly less than that of the control, but both of the differential buds showed albino. Finally, 3.5 mg/L concentration of glufosinate-ammonium were determined as criterion of selection pressure.
 Figure 3 Effect of glufosinate-ammonium on the clustering shoot regeneration of soybean cotyledonary-nodes of Hajiao5337 and Hajiao5489 |
1.3 Target gene of T1 generation plant validated
Plasmid pCAMBIA3300-HSF8 was used as a positive control, non-transgenic regenerated plants and Hajiao5337 and Hajiao5489 as references, sterile water instead of template DNA as a negative control, the total DNA of the resistant plants were used as DNA template to amplify specific bar gene by using 35s-bar of primers. The results shown in Figure 4, indicated that 17 resistant plants were exhibited positive, which preliminary proved hsf8 gene to be integrated into the soybean genome.
|
Figure 4 The PCR analysis of genomic DNA in putative transgenic soybean plants T1 generation Note: M: DL2000; 1: Positive control of pCAMBIA3300-HSF8; 2: Negative control of distill deionized water; 3~13: DNA of T1 generation from independently transformed plants; 14: DNA from untransformed plant Hajiao5337 (negative control); 15: DNA from untransformed plant Hajiao5489 (negative control) |
1.4 hsf8 expression level of T1 generation plant detected by Real-time PCR
The expression of hsf8 gene in T1 generation of transgenic plants derived from Hajiao5337 and Hajiao5489 as receptor were detected by Real-time PCR, data shown in Table 1 and Table 2. △△CT standed for different CT value between the target gene hsf8 and reference gene (lectin). The expression level of hsf8 gene among tested plants were compared based on differential expression in multiples (2-△△CT), that is the ratio come from initial concentration of hsf8 gene in transgenic plants to that of non-transgenic plants. Nine individuals were exhibited significant increases of gene expression, three of them were increased twenty times than that of non-transgenic plants, three plants for fifteen times increase and two plants for five times increase (Figure 5). Compared with the expression of Hajiao5489, the individual, T07Ⅱ-002-2, exhibited obvious differences in gene expression level that was 48 times less than the amount of gene expression of Hajiao 5489 (Figure 6).
.png) Table 1 Expression level of the T1 generation transgenic hsf8 plants derived from Hajiao5337 identified by Real-time PCR |
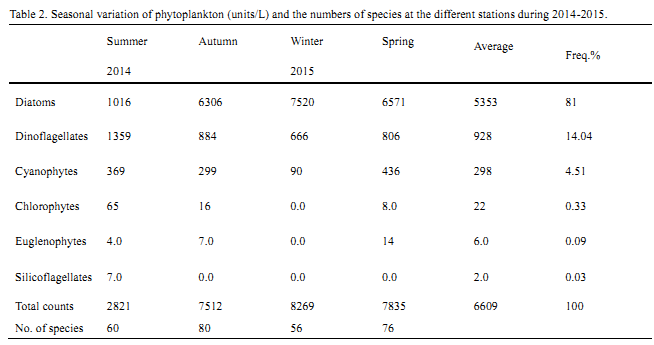 Table 2 Expression level of the T1 generation transgenic hsf8 plants derived from Hajiao5489 identified by real-time PCR |
 Figure 5 hsf8 expressions of individual T07-â… transgenic plants of T1 generation and non-transgenic plants Hajiao5337 by Real-time PCR analyses Note: 1: Hajiao5337; 2: T07â… -003-1; 3: T07â… -003-2; 4: T07 â… -005-1; 5: T07â… -002-2; 6: T07â… -006-1; 7: T07â… -006-2; 8: T07â… -006-3; 9: T07â… -007-1; 10: T07â… -007-2; 11: T07â… -007-3; 12: T07â… -010-1; 13: T07â… -010-2; 14: T07â… -010-3; 15: T07â… -014; 16: T07â… -015-1; 17: T07â… -015-2; 18: T07â… -015-3 |
 Figure 6 hsf8 expressions of individual T07â…¡ transgenic plants of T1 generation and non-transgenic plants Hajiao5489 by Real-time PCR analyses Note: 1: Hajiao5489; 2: T07â…¡-002-1; 3: T07â…¡-002-2; 4: T07â…¡-003; 5: T07â…¡-010-1; 6: T07â…¡-010-2 |
Through the above results, the preliminary results indicated that hsf8 gene expression variations in transgenic plants should be due to insertion of the target gene hsf8.
2 Discussions
Soybean genetic transformation was considered as one of the most difficult conundrums in soybean research, to date soybean transformation system is still inefficient and poor reproducibility. Increasing transformation efficiency depends on the establishment of perfect regeneration system, genetic transformation system and smart screening strategy (Li et al., 2006; Paz et al., 2006). In this paper, we adopted a modified screening method, which recovery culture was carried out prior to co-culture procedure. The results we obtained were the similar with the previously reported publication (Liu et al., 2007). It is noteworthy that recovery culture time might be too long to inhibit the growth of transformed cells due to excessive proliferation and differentiation of non-transformed cells, which may be inconducived to transformation efficiency. Bar gene as a selective marker gene is less restricted to receptor’s genotypes, which could be used for transformation in almost all species, in this point it would be helpful to improve the genetic transformation efficiency.
Many studies indicated that there were significant differences in the capacities of Agrobacterium infection in different plants. Therefore, the match degree between the Agrobacterium and plant would be very important in the research of genetic transformation, in other words, ideal combination between Agrobacterium and plants would directly affect transformation efficiency. In this study, the two kinds of Agrobacterium GV3101 and LBA4404 were chosen for transformation of two soybean lines, both of two strains had a strong ability to infect soybean cotyledonary-node to generate much more resistant buds. In comparison, the infective activity of strain GV3101 was better than that of LBA4404. However, it was difficult to be sterilization after co-culture due to its high infective activity, which resulted in acquiring resistant seedlings. We could not remove out all Agrobacterium even if the concentration of cephalosporin reached to 600 mg/L. It is obvious that the growth of resistant buds will be influenced if increasing the usage of antimicrobial. So choosing antimicrobial is also important as choosing the active bacterial strain.
Northern blot method and RT-PCR quantitative method are commonly used in gene expression research. However, the Real-time PCR would be more convenient, faster and more accurate than that of Northern blot and RT-PCR to detect the abundance of gene expression in a variety of tissue cells, to analyze gene expression and regulation, to monitor mRNA expression mode, to detect the presence of genes in a small amount of tissue, to track the clone in the cell population and to quantitatively analyze gene transcription levels in different tissues (Livak and Schmittgen, 2001).
In this study, we conducted hsf8 expression analysis of transgenic plants and non-transgenic plants by using real-time PCR. Because introduced gene come from soybean self, the non-transgenic soybean lines, Hajiao5337 and hajiao5489, exhibited a reasonable level of gene expression. In this research, there are evidences of over-expression and low expression in transgenic plants, the reasons might be increase of gene copy numbers to enhance the level of expression, whereas gene silencing might be the reason for low expression in some transgenic plants.
3 Materials and Methods
3.1 Plant materials
Two high yield soybean new lines, Hajiao5337 and Hajiao5439, developed by our research group, used to be transformation receptors in this research.
3.2 Strains and plasmids
Plasmid pBI121-HSF8 containing hsf8 gene and plant expression vector pCAMBIA3300 were provided by Dr Baoge Zhu from IGDB of CAS. E. coli strain DH 5 was bought from TIANGEN Company and Agrobacterium tumefaciens strains GV3101 was deposited in this lab.
3.3 Chemicals and reagents
Taq DNA Polymerase and T4 DNA ligase were purchased from TaKa-Ra Company; restriction enzymes from Promega Corporation; Glufosinate-ammonium, 6-Benzylaminopurine (6-BA) and Indole-3-utyric (IBA) from Sigma company. RNAprep Plant Kit from TIANGEN Company, Power SYBR® Green PCR Master Mix and MicroAmpTM Optical Adhesive Film Kit were purchased from ABI Research company, and reverse transcription reagents were purchased from Invitrogen. And sequencing primers were synthesized by Shanghai Sangon. Antibiotics, kanamycin and cephalosporin, were domestically produced with analytical grade in China.
3.4 hsf8 expression vector construction based pCAMBIA3300
Single colony, pBI121-HSF8 or pCAMBIA3300 was picked to be cultured at 200 r/min shaking incubation in LB liquid medium (Kan+) at 37℃ until the OD600 value equal 0. 4 measured. Plasmid pBI121-HSF8 was digested with EcoRâ… and Hind â…¢ restriction enzyme, then about 2.8 kb fragments were recovered, which contained the completely 35S + hsf8 + NOS expression structure. While plasmid pCAMBIA3300 was digested with EcoRâ… and Hind â…¢ restriction enzyme, then about 8.5 kb fragments were recovered. Two recovered fragments were ligated by T4 ligase overnight at 22℃. The connected products were transformed into E. coli DH5 competent cells by heat shock method. Positive recombinants were screened by using kanamycin (Km) 100 mg/L, then positive plasmids were transformed into Agrobacterium competent cells, and coated on plate of YEB (Kan +, Rif + and Str +) for 48 h dark incubation until strain plaque growing about 2 mm diameters.
3.5 Agrobacterium mediated transformation
3.5.1 Screening concentration of glufosinate-ammonium
According to preliminary results, delay screening method was employed in this research, The soybean cotyledonary-node was applied to bud induction culture medium without selecting agents about 3 days until the differential bud spots on cotyledonary-node appeared. Then the visible buds of cotyledonary-node were transferred into bud induction media with different concentrations of glufosinate-ammonium for screening culture. Concentrations of glufosinate-ammonium were designed as 0 mg/L, 0.5 mg/L, 1 mg/L, 1.5 mg/L, 2 mg/L, 2.5 mg/L, 3 mg/L, 3.5 mg/L and 4 mg/L. Thirty explants were placed on media of each concentration treatment with a duplicates and were incubated for two weeks at (26±1)℃. The differential rate of cotyledonary-node was calculated following as the differential cotyledonary-node number by total number of cotyledonary-node incubated.
3.5.2 Genetic transformation
Preparation of cotyledonary-node: The selected soybean seeds were disinfected following the method of chlorine disinfection (Liu and Wei, 2002). The sterilized soybean seeds placed in the germination medium (1/2 MSB (half inorganic salt ingredients of MS medium + half organic ingredients of B5 medium), 0.7% agar, pH 5.8). Taking 5~6 day cultured asepsis seedlings, and cutting cotyledonary-node from vertical direction to retain 3~4 mm hypocotyl as well as removing the terminal bud and lateral bud then placing into the pre-culture medium (B5 medium +1.7 mg/L 6-BA +0.1 mg/L IBA, 0.7% agar, pH 5.7).
Strains Preparation: Picking up single plaques of Agrobacterium GV3101 with pCAMBIA3300-HSF8 and LBA4404 from culture plates, Being Inoculated with YEB liquid medium containing antibiotics (40 mg/L Rif, 50 mg/L Str and 100 mg/L Kan) at 200 r/min shaking culture for 12~24 h at 28 ℃ until OD600 value going to about 0.6. Supernatant was removed after 10 min centrifugation at 4 000 r/min, then the sediment was resuspended with the same volume of YEB ready for use.
Transformation and screening culture: Cotyledonary-nodes with pre-cultured 1 d were placed into the Agrobacterium strain liquid for infection about 25~30 min, then succeed to co-culture medium (B5 medium +1.7 mg/L 6-BA +0.1 mg/L IBA +100 mg/L ferulic acid, 0.7% agar, pH 5.2), cultured in the dark for 3~4 d. The infected cotyledonary-nodes were rinsed by sterile water containing 500 mg/L Cef by 4~5 times, then being inoculated into the sterilized medium (B5 +1.7 mg/L 6-BA +0.1 mg/L IBA +600 mg/L Cef, 0.7% agar powder, pH 5.7) for a week until he buds were appearing. The explants with buds were transferred into selective medium (B5 +1.7 mg/L 6-BA +0.1 mg/L IBA +600 mg/L Cef +3.5 mg/L glufosinate-ammonium, + 0.7% agar, pH 5.7) (Tang et al., 2008).
Growth and rooting of the plants with resistance: The buds were placed on the growth medium (B5 +1.7 mg/L 6-BA +0.1 mg/L IBA +600 mg/L Cef +1.75 mg/L glufosinate-ammonium, 0.7% agar, pH 5.7) after two weeks screening. Cutting the base of shoots for immersing 1 minute on the filted IBA(1 mg/L) once the clustering buds grew 3~4 cm in length, and then immediately placed the treated shoots into rooting medium MSB, the plantlets were transplanted in the flower pots until the root system well developed with a lateral root appearing.
3.6 Transgenic plants detected by PCR
Using leaves of resistant plants screened by glufosinate-ammonium to extract soybean genomic DNA by CTAB. Designing a primer that included the F: 5'-TTCGCAAGACCCTTCCTC-3' and R : 5'-ACCCACGTCATGCCAGTT-3' based on CaMV35S promoter and bar gene sequence for PCR amplification, which was synthesized on the ABI 9700 by Shanghai Sangon Bioengineering Co., Ltd. Amplified fragment was expected to 594 bp in length. PCR reaction were performed following the protocols as 94℃ for pre-denaturation 5 min in advance, then 30 cycles with 94℃ for denaturation 30 s, 55℃ for annealing 30 s, 72℃ for extension 30 s, and finally 72℃ for extension 5 min.
3.7 T1 generation transgenic resistant plants detected by Real-time PCR
The three-week-old seedlings of T1 generation transgenic plant and non-transgenic reference plants were transferred into the greenhouse culture at the room temperature, and then total RNAs were extracted from leaves of transgenic hsf8 plants and non-transgenic plants, respectively. Total RNAs were reverse transcripted to cDNA. Using soybean lectin gene as an internal reference, two pairs of primers were designed based on internal reference hsf8 gene, one pair of primers including F: 5'-ATAACTCGGCGGAAACCT-3', R: 5'-TTGCTGTCGTTGCTCCAT-3' was for detecting hsf8 gene. And another pair of primers was including F: 5'-CTTCGCCGCTTCCTTCAA-3', R: 5'-GCCCATCTGCAAGCCTTTT-3' for detecting lectin gene. Real-time PCR reaction was performed on the real-time quantitative fluorescene PCR instrument Mx3000pTM. Reaction conditions were following as: 95℃ for denaturation at 10 min in advance and then 40 cycles as 95℃ for denaturation 30 s, 60℃ annealing 1 min.
Housekeeping gene was set up as Normal, leaf cDNAs of Hajiao 5337 and hajiao 5489 as Calibrators, and leaf cDNAs of T07â… and T07â…¡, the lines of T1 generation plants, as unknown, repeated thrice , setting up one for both the target gene and housekeeping gene NTC. Drawing amplification curve and melting curve. Quantitative results were placed on the coordinate map by taking the average of 3 times repeats. The relative expression was calculated by using comparative CT method (△△CT) that was, the relative expression =2-△△CT=2-(â–³CT sample -â–³CT control)= 2-[(CT sample-CT internal reference) - (CT control -CT internal reference)].
Authors' contributions
XT, MG, ZY and GP are the persons who designed and conducted this experiment; LL, LZ and LW finished the data analysis and paper preparation. LL is the PI of this project involving in project design, data analysis, and paper modification. All authors had read and consented the final text.
Acknowledgements
This research is jointly sponsored by the National 863 Project (No. 2006AA1021F9) and National Special Transgene Projects (No. 2008ZX08004-002). Authors thank for two anonymous reviewers with their critical comments. In this paper we mentioned some chemical and reagent suppliers and sequencing service providers, that doesn't mean we would like to recommend or endorse the production of theirs.
References
Busch W., Wunderlich M., and Schöffl F., 2005, Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana, Plant J., 41(1): 1-14 http://dx.doi.org/10.1111/j.1365-313X.2004.02272.x PMid:15610345
Lee J.H., and Schöffl F., 1996, An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana, Mol. Gen. Gent., 252(1-2): 11-19 http://dx.doi.org/10.1007/s004389670002 http://dx.doi.org/10.1007/BF02173200
Lee J.H., Hübel A., and Schöffl F., 1995, Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermo tolerance in transgenic Arabidopsis, Plant J., 8(4): 603-612 http://dx.doi.org/10.1046/j.1365-313X.1995.8040603.x
Li M.C., Cai Y., Zhao G.L., Caiyin Q.G.L., Zhou H., Sun W., and Xing L.J., 2006, Improvement of cotyledon node regeneration system in soybean (Glycine max), Zuowu Xuebao (Acta Agronomica Sinica), 32(2): 223-227
Liu H.K., and Wei Z.M., 2002, A method for sterilizing mature seeds of soybean, Zhiwu Shenglixue Tongxun (Plant Physiology Communications), 38(3): 260-261
Liu S.J., Huang J.Q., and Wei Z.M., 2007, Factors influencing Agrobacterium-mediated cotyledonary-node transformation of soybean (Glycine max L.), Fenzi Xibao Shengwu Xuebao (Journal of Molecular Cell Biology), 40(5): 286-294
Livak K.J., and Schmittgen T.D., 2001, Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-△△C(T)) method, Methods, 25(4): 402-408 http://dx.doi.org/10.1006/meth.2001.1262 PMid:11846609
Paz M.M., Martinez J.C., Kalvig A.B., Fonger T.M., and Wang K., 2006, Improved cotyledonary node methond using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation, Plant Cell Rep., 25(3): 206-213 http://dx.doi.org/10.1007/s00299-005-0048-7 PMid:16249869 http://dx.doi.org/10.1007/s00299-005-0113-2
Tang X.F., Liu L.J., Zhang X.M., Xue Y.G., Yang Z., Gao M.J., Zhang L., and Wei L., 2008, Improvement of regeneration system in high-yield soybean lines Hajiao5337 and Hajiao 5489, Dadou Kexue (Soybean Science), 27(2): 203-207
Wang W.X., Vinocur B., and Altman A., 2003, Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance, Planta, 218(1): 1-14 http://dx.doi.org/10.1007/s00425-003-1105-5 PMid:14513379
Zhu B.G., Ye C.J., Lv H.Y., Chen X.J., Chai G.H., Chen J.N., and Wang C., 2006, Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max), J. Plant Res., 119(3): 247-256 http://dx.doi.org/10.1007/s10265-006-0267-1 PMid:16570125
. PDF(1744KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaofei Tang
. Lijun Liu
. Mingjie Gao
. Zhe Yang
. Guofeng Pu
. Lei Zhang
. Lai Wei
Related articles
. Soybean ( Glycine max )
. Heat shock factor 8 gene ( hsf8 )
. Agrobacterium -mediated transformation
. Heat tolerance
Tools
. Email to a friend
. Post a comment


