2 School of Biology and Basic Medical Sciences, Soochow University, Suzhou, 215123, R.P. China
 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2012, Vol. 3, No. 4 doi: 10.5376/tgg.2012.03.0004
Received: 16 Oct., 2012 Accepted: 29 Oct., 2012 Published: 12 Nov., 2012
Cai and Liu, 2012, Characrerizing the Sequences of Kr Gene in Different Genotypes of Common Wheat, Triticeae Genomics and Genetics, Vol.3, No.4 38-43 (doi: 10.5376/tgg.2012.03.0004)
The incompatibility of distant hybridization in wheat has been known to be controlled by the Kr gene. In order to figure out the characteristics of the Kr gene in different genotypes of wheat, the six genotype germplasms selected from the Chinese wheat micro-core collection, called Mazhamai, Xiaobaimang, Tuokexunyihao, Chinese spring, Yangmai, and Zhengmai9023, were employed to be experimental materials for identifying the characteristic of the Kr genes by means of the homology cloning approach. The results showed that the three of six genotypes could amplify a fragment of Kr gene with 414 bp in length. The sequences among three cloned fragments shared 99.8% homology except two sites with SNP differences. Blast analysis indicated that the cloned sequences of partial Kr genes have 85% homology with the S-locus receptor kinase gene in the same regions. Thus, we preliminarily speculated the Kr gene might possess similar functions as the gene conferring self-incompatibility in plant,which possibly facilitated cellulae mastoideaes to accumulate calloses in chapitersand affected the competitive ability of exogenous pollens eventually resulting in the incompatibility of distant hybridization in wheat.
The relative genera and species of common Wheat (Triticum estivum L.) bear a lot of excellent genes. By means of distant hybridization the favorable genes of relative plants could be transferred into wheat (Dao, 2009), which would be important significance of enriching wheat genetic basis and breeding excellent new variety. However, the most important issue facing the wheat distant hybridization is the genes, the dominant Kr1 and Kr2 genes (commonly call Kr gene), located in chromosomes of 5BL and 5AL chromosome, which can effectively inhibit the heterologous pollen mating with the host to decrease hybridization affinity. So, the Kr gene also was known as the gene of distant hybridization incompatibility in wheat (Manickavelu et al., 2009b).
The studies showed that the activity of the Kr gene in the different system of the distant hybridization were very different, In the hybridization in the closer relative species e.g. wheat×rye (Secale cereale L.), the dominant Kr gene actively suppresses the pollen affinity, whereas in distantly relative species e.g. wheat×maize (Zea mays L.), the dominant Kr gene maintains inactive (Torres et al., 2010). So far, what the mechanism of the Kr gene suppresses the distant hybridization is unclear yet.
Previously, we happened that there were significant differences of hybridization affinity in distant hybridization of wheat and maize, either in the crosses between the same genotype wheat and different corns or in the crosses of different genotype wheat and the same genotype corn. Furthermore, by means of the treatments of low temperature, dark culture, different concentrations of 2,4-D and processing times, as well as corn pollen repeated pollination etc., the differences of hybridization affinity also existed significantly (Cai et al., 2006; 2007). These phenomena implied that the Kr gene should possess rich allele diversities and complex expression and regulation.
Recently, Manickavelu obtained a 751 bp cDNA sequence of Kr gene from the recombinant inbred lines RIL (Kr/kr) derived from the cross of Chinese Spring 5B monomer (kr) and Mara (Kr/Kr) by using the Annealing Control Primer System, (ACP) (GenBank accession No.: AB379558.1, due to the stop codon existing in the sequence, therefore assigned as pseudo-gene mRNA) (Manickavelu et al., 2009a). So far, it was not yet to be reported about the molecular structure of Kr gene in different wheat varieties or germplasms.
For the above reasons, the six genotypes of common wheat selected from 285 Chinese wheat core germplasms were employed to isolate the Kr gene by using homologous cloning approach with the Kr gene-specific primers, in order to predict the encoding proteins of the Kr genes and their functions and to provide a theoretical basis for explaining the molecular mechanism of incompatibility of distant hybridization in wheat and enhancing the distant hybridization efficiency, promoting genetic improvement and germplasm innovation in wheat.
1 Results and Analysis
1.1 PCR amplification of the fragments of Kr gene
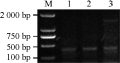
PCR amplification was carried out with specific primers L4 under the same conditions. The results showed that one expected PCR band appeared in three genotypes of Mazhamai Xiaobaimang and Tuokexun- yihao, and the sequencing results exhibited that the length of three bands was 414 bp (Figure 1).
Figure 1 The fragment of Kr gene amplified by PCR with primer L4
1.2 Alignment of the sequences of Kr gene fragments
PCR products were recovered and purified as well as TA cloned and sequenced. Alignment of the sequenced target products were conducted by applying DNAMAN Software. The results of sequence homology comparison showed that the sequence fragment homology of the Mazhamai and Xiaobaim- ang shared 100% identity, while 99.8% with Tuoke- xunyihao that only existed two SNP differenced at the sites of 284 bp and 321 bp (Figure 2), suggesting that the sequences of this fragment in the Kr gene should be highly conservative in different wheat genotypes.
Figure 2 Sequence alignment of Kr gene fragments in three genotypes wheat

1.3 Homologous sequences and their relationships of Kr gene sequences in Triticease plants
Thirteen sequences of Triticeae plants with high homology (E value <1e-20) were blasted at NCBI by using the fragment sequence of Kr gene obtained in this research as probe. The obtained sequences included the S-locus receptor kinase gene (AY963808. 1), high molecular weight glutenin gene (AY368673.1), CRT/DRE binding factor (AY951949.1), EXPB1 gene (AY533103.1). We applied these sequences to construct the cluster based on the Distance tree of results program of NCBI website (Figure 3).
Figure 3 Kr genes homologous sequences and their relationship in Triticeae plants

Comparing the obtained 414 bp fragment of Kr gene with wheat S locus (S-locus) receptor kinase gene (AY963808.1), the blast results showed that the sequence homology reached 85% in the same regions (Figure 4), indicated that the encoding proteins of the obtained sequences might share a similar function with S-locus receptor kinase in plant.
Figure 4 Sequence alignment between the obtained fragment of Kr gene and S-locus receptor kinase gene
![]()
2 Discussions
DNA sequence of the Kr gene wheat has not been reported so far. Only 14 expression sequence tag (EST) and a cDNA sequence of Kr gene in wheat could be retrieved in GenBank, which seemed no overlapping and unknown nucleotides by detection in DNAstar software with the trimmed optimization of 14 ESTs, and furthermore these sequences could not carry out the sequence assembly used for a template designing primer.
Therefore, in this research, the only cDNA sequence (GenBank accession No.: AB379558.1) was employed to design primers. However the primer designed based cDNA sequences applied to amplified genomic DNA that might result in leaping over the RNA splice sites of primers due to the presence of the intron as well as being difficult to specifically match the template resulting in amplification failure. This would be one of the reasons why only three of the six chosen materials yielded target products.
Obviously, the activity of the Kr gene exhibited differences in distant hybridization system, The pollination between wheat and rye was more difficult than the of wheat and maize, which indicated that the diversity of the Kr genes and the complexity of the Kr gene expression should be existence. Although the Kr gene sequence obtained in the three genotypes shared high homology in this research, we failed to obtain the Kr gene in another three genotypes. So we speculated that there would be more plenty polymorphisms of Kr gene sequence happening in large testing samples.
Studies have shown that self-incompatibility S locus (S-locus) is the multiple alleles encoding S locus receptor kinase (SRK) (Stein et al, 1991), S-locus glycoprotein (SLG) (Nou et al., 1993) and S-locus cysteine-rich protein (SCR) (Schopfer et al., 1999), wherein, SCR is signaling molecules of self-incompatibility (SI), specifically expressed in pollen that plays the role of ligand; SLG and SRK express in the pistil mastoid where SRK is the recognition factor for SI signal.in the stigma acting as the receptor but SLG act as assistant receptor. Interaction caused by pollen ligand receptor and stigmatic papillae cells results in SI signal transduction in the stigma papillae, eventually leading to the reaction of self-incompatibility (Takayama and Isogai, 2005;Fujimoto and Nishio, 2007; Zhang et al., 2010). In this research the obtained Kr gene fragment shared 85% homology with wheat S locus receptor kinase gene within the same region, which would provide an insight to further understand the function of the Kr gene in wheat.
3 Materials and Methods
3.1 Experimental materials
Six genotypes were chosen from the 285 Chinese wheat mini-core germplasms as the experimental materials according to the report of Qi etc. (1997) and our previous results of the hybridization between wheat and corn (Cai et al., 2006; 2007), the selected germplasms were Mazhamai, Xiaobaimang, Tokesun- yihao, Chinese Spring, Yangmai, and Zhengmai9023, The seeds germinated one week at 25℃, young leaves were picked to extract genomic DNA.
3.2 Extraction of genomic DNA
Genomic DNA was extracted followed with the instructions of the kit for extraction plant genomic DNA developed by the Transgen company (http://www.transgen.com.cn/uploadfile/201111/20111111093152454.pdf).
3.3 Primer design and PCR amplification
A pair of primers was designed based on the cDNA sequence of Kr gene (GenBank accession No.: AB379558.1) as a template by using the software of Primer 5.0, the sequence of Forward Primer 5'-TCCGCGCGCCCCTAGAGAAA-3', and the Reverse Primer 5'-TCGCCTCCACGCTTCACCCT-3', which synthesized by Shanghai Sango Biological Engineering Company.
The 50 μL PCR reaction system included as follows as: 5 μL of 10×PCR buffer (adding Mg2+), 4 μL of 2.5 mmol/L dNTPs, 1 μL of 1 μmol/L Forward Primer and Reverse Primer each, 0.5 μL of 5 U/μL DNA Polymerase, and 50 ng of the DNA template, finally by adding sterilized ultrapure water up to 50 μL. The above chemicals were purchased from Beijing Transgen Company.
PCR procedures were set as follows: pre-denaturation at 94℃ for 5 min. and then 38 cycles with the process following denaturation at 94℃for 45 s, annealing at 54℃for 45 s, and extension at 72℃ for 1.5 min; final extension at 72℃ for 10 min. The PCR products were detected by electrophoresis with 1% agarose gel (containing ethidium bromide), with the conditions of 1×TAE electrophoresis buffer solution, 120 V voltage and running 25 min, the staining gel was observed under the imaging scanner and deposited in computer. PCR amplification product was purified by the Shanghai Sango Biological Engineering Company to do TA cloning prior to sequencing.
3.4 Sequence Alignment
Alignment of sequence homology comparison for the obtained sequences was carried out by using DNAMAN software; the high homology sequences (E value <1e-20) were searched using the obtained Kr gene sequence as a probe at the NCBI database.
Author contributions
CH and LQQ are the executors for the experimental design and lab work in the study to complete data analysis, manuscript writing and revising; CH conceived this project and took in charge. Both authors read and approved the final manuscript.
Acknowledgements
This study were jointly funded by the Open Foundation by the Key Laboratory of Crop Genetics and Germplasm Enhance- ment (ZW2010002) and the Major Project of the 12th Five-Year National Science and Technology Support Program entitled Technology Engineering for Grain Production (2011BAD16- B06 and 2012BAD04B09).
References
Cai H., Ma C.X., and Lu W.Z., 2006, Wheat haploid plant produced by embryo culture of Wheat×Maize, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 14(4): 633-634
Cai H., Ma C.X., Qiao Y.Q., and Lu W.Z., 2007, Establishment of regeneration system in wheat haploid plant through wheat×maize, Jiguang Shengwu Xuebao (Acta Laser Biology Sinica), 16(1): 43-48
Dao W.W., 2009, Wide hybridization: engineering the next leap in wheat yield, J. Genet. Genomics, 36(9): 509-510
Fujimoto R., and Nishio T., 2007, Self-incompatibility, Advances in Botanical Research, 45: 139-154
Manickavelu A., Koba T., Mishina K., and Sassa H., 2009a, Identification of differential gene expression for Kr1 gene in bread wheat using annealing control primer system, Mol. Biol. Rep., 36: 2111-2118
Manickavelu A., Koba T., Mishina K., and Sassa H., 2009b, Molecular characterization of crossability gene Kr for intergeneric hybridization in Triticum aestivum (Poaceae: Triticeae), Plant Systematics and Evolution, 278(1-2): 125-131
Nou S., Watanabe M., Isogai A., and Hinata K., 1993, Comparison of S-alleles and S-glycoproteins between two wild populations of Brassica campestris in Turkey and Japan, Sexual Plant Reproduction, 6(2): 79-86
Qi Z.J., Li Q.Q., and Wang H.G., 1999, Crossability of some land wheat races from Shandong with rye, Huabei Nongxuebao (Acta Agriculturae Boreali-Sinica), 14(2): 15-1
Schopfer C.R., Nasrallah M.E., and Nasrallah J.B., 1999, The male determinant of self-incompatibility in Brassica, Science, 286: 1697-1700
Stein J.C., Howlett B., Boyes D.C., Nasrallah M.E., and Nasrallah J.B., 1991, Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea, Proc. Natl. Acad. Sci. USA, 88(19): 8816-8820
Takayama S., and Isogai A., 2005, Self-incompatibility in plants, Annu. Rev. Plant Biol, 56: 467-489
Torres L.E., Bima P., Maich R., Badiali O.J.J., and Nisi J., 2010, Production of haploid plants from ten hybrids of bread wheat (Triticum aestivum L.) through wide hybridization with maize (Zea mays L.), Agriscientia, 2: 79-85
Zhang L., Zhu L.Q., Jia H., Li M., Wu W.R., Tang Z.L., and Wang X.J., 2010, Methylation Analysis of S-locus receptor kinase gene (SRK) coding region of Brassica oleracea, Nongye Shengwu Jishu Xuebao (Journal of Agricultural Biotechnology), 18(3): 482-488
. PDF(351KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Hua Cai
. Qingqing Liu
Related articles
. Wheat
. Kr gene
. Distant hybridization
. Plant self-incompatibility
Tools
. Email to a friend
. Post a comment


