Identification of AhAQ1 Encoding a Putative Aquaporin whose Expression is Regulated by Salt Stress In Peanut (Arachis hypogaea L.) 
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2010, Vol. 1, No. 5 doi: 10.5376/lgg.2010.01.0005
Received: 21 Jun., 2010 Accepted: 11 Aug., 2010 Published: 18 Oct., 2010
Pan et al., 2009, Identification of AhAQ1 Encoding a Putative Aquaporin whose Expression is Regulated by Salt Stress In Peanut (Arachis hypogaea L.), Molecular Plant Breeding, 7(5): 867-872 (doi: 10.3969/mpb.007.000867)
The primary role of aquaporins is to control water transport in plants, which might function in plant response against salt stress. In this research, we obtained the full-length cDNA of an aquaporin gene AhAQ1 by RT-PCR approach from salt-stressed peanut leaves. AhAQ1 encodes a protein consisting of 287 amino acids with the calculated molecular mass of 30.57 KD and the isoelectric point of 9.04. Sequence analysis indicated that AhAQ1 contained six transmembrane domains and two NPA-boxes and phylogenetic analysis showed that the AhAQ1 clusters were the same with the PIP1 family. The expression pattern of AhAQ1 in peanut tissues under high-salt treatment was examined by semi-quantitative RT-PCR, The results showed that the accumulation of AhAQ1 transcripts was induced by salt stress . The combination of these results illustrate that AhAQ1 may play a role in peanut in response to salt stress.
Soil salinity is one of the major stresses in the world that affects plant growth and causes a significant loss of crop productivity (Wang et al., 2003; Zhu, 2003). The high concentration of sodium ions induceion toxicity and osmotic stress by limiting absorption of water from soil. Aquaporins represent a large family with highly conserved sequence in plants (Johanson et al., 2001; Chaumont et al., 2001; Sakurai et al., 2005). Previous studies confirmed that aquaporins play important roles in plant water transport by heterologous expression in Xenopus laevis oocytes (Maurel et al., 1993; Preston et al., 1992; van Hoek and Verkman et al., 1992). They form proteinaceous pores that facilitate the passive diffusion of water across membranes of living cell by increasing the osmotic hydraulic conductivity of the membrane (Preston et al., 1992; Siefritz et al., 2002). Based on their sequence similarities or structural features, plant aquaporins may be divided into four groups: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin26-like intrinsic proteins (NIPs) and small basic intrinsic proteins (SIPs) (Johanson et al., 2001; Johanson and Gustavsson 2002; Chaumont et al., 2005; Murata et al., 2000), Among the four categories, the plasma membrane intrinsic proteins (PIPs) ,which are further classified into two subfamilies, PIP1s and PIP2s (Luu and Maurel, 2005), according to the amino acid residues at the N- and C-terminals and the conserved NPA box.
Since the first plant aquaporin γ-TIP was isolated from Arabidopsis (Maurel et al., 1993), many other aquaporins have been identified from plants involving Arabidopsis, maize, tobacco and rice, and so on (Johanson et al., 2001; Chaumont et al., 2001; Mahdieh et al., 2008; Sakurai et al., 2005). Numerous studies confirmed that many plant aquaporin genes are involved in the response to water stresses such as drought and salt. For example, the amount of rice water channel protein RWC3 increased in upland rice under drought stress, whereas decreased in lowland rice, and overexpression of RWC3 in lowland rice could result in an enhanced tolerance to drought stress (Lian et al., 2004). Another instance is that transgenic tobacco plants expressing sense BnPIP1 gene showed an increased tolerance to water stress (Yu et al., 2005). Three genes named NeMip1, NeMip2 and NeMip3 have been isolated from tobacco, and their mRNA were accumulated by salt and drought stress (Yamada et al., 1997). Besides,the expression of barley HvPIP2;1 was downregulated after high salt treatment, and constitutive expression of HvPIP2;1 raised salt sensitivity of transgenic rice (Katsuhara et al., 2003).
Peanut is one of the major oilseed crops in the world, and the increase of the soil salinity might heavily limit its yield. To study the molecular mechanisms of peanut to salt tolerance, in this research, we isolated several salt-inducible genes from the salt tolerant variety Huayu28, and then one of them, an aquaporin gene, was chosen for further characterization.
1Results
1.1 Cloning and characterization of AhAQ1 gene
The full length of AhAQ1 gene was cloned from salt-treated peanut leaves by RT-PCR technology. AhAQ1 encodes a protein consisting of 287 amino acids with the calculated molecular mass of 30.57 KD and the isoelectric point of 9.04 (Figure 1). Sequence analysis of deduced amino acids indicated that AhAQ1 was an highly hydrophobic with 6 possible transmembrane domains linked by loop A-E, and two NPA boxes, which can be found in most aquaporins that lies in loop B and loop E (Figure 2). A comparison used the deduced amino acids of AhAQ1 as a query probe to search gene database (http://www.ncbi.nlm.nih.gov) though blastp algorithm program showed that AhAQ1 was more similar to the PIP2 family. Multiple sequence alignment showed that the amino acid sequence of AhAQ1 was highly conserved in other PIP2s from maize, barley, tobacco, Arabidopsis and olive, especially in the transmembrane domains (Figure 3).
 Figure 1 Nucleotide sequences and deduced amino acid residues of AhAQ1 Note: The amino acid residues are indicated by a single letter code; The transmembrane domains and NPA motifs are underlined and double-underlined, respectively |
 Figure 2 Hydrophobic plot of AhAQ1 Note: The hydrophobic values were calculated by the program TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html); The amino acid residues 1~287 are presented at X-axis. Areas above the x-axis indicate hydrophobic portions of the protein, and that under the line are hydrophilic ones. It shows that there are at least 6 possible membrane-spanning regions in the AhAQ1; 1~6: 6 transmembrane domains; A,B: NPA box |
 Figure 3 Alignment of AhAQ1 with PIP2-type proteins in other plants Note: Positions containing identical residues are shaded in black, while conservative residues in grey; The transmembrane domains and NPA motif are lined; The alignment was performed using ClustalX (ver 1.81) program and viewed by GeneDOC software |
To investigate the evolutionary relationships among plant aquaporins, a phylogenetic tree was constructed using a neighbor-joining method with the full-length amino acid residues (Figure 4). The result showed that AhAQ1 was grouped into the big branch of PIP2-type proteins, which including AtPIP2;3, ZmPIP2;1, HvPIP2;1, OePIP2;1, BoPIP3 and NtPIP2;1. On the other hand, PIP1 proteins including NtPIP1;1, AtPIP1;1, BoPIP1b1, OePIP1;1, ZmPIP1;2 and HvPIP1;1, which have longer N-terminal extensions and shorter C-terminal ends compared with the PIP2 proteins (Schaffner, 1998), were categorized into another big branch.
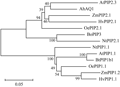 Figure 4 Phylogenetic tree analysis of AhAQ1 and PIP-type proteins in other plants Note: The tree was constructed by neighbor-joining method with MEGA (ver 3.1) program; Branch numbers represent percentage of bootstrap values in 1000 sampling replicates and the scale indicates branch length; The corresponding proteins are listed as follows: maize, ZmPIP1.2 (AAD29676), ZmPIP2.1 (AAK26758), barley, HvPIP1.1 (BAF41978), HvPIP2.1 (BAA23744), Arabidopsis, AtPIP1.1 (X75881), AtPIP2.3 (D13254), Brassica, BoPIP1b1 (AAG23179), BoPIP3 (AAG30607), olive, OePIP1.1 (DQ202708), Oe-PIP2.1 (DQ202709), tobacco, NtPIP1.1 (AF440271), NtPIP2.1 (AF440272) |
1.2 Expression of AhAQ1 in response to high salt stress
The expression pattern of AhAQ1 in peanut tissues under high salt treatment was examined by semi-quantitative RT-PCR. The results showed that the transcripts of AhAQ1 were responsive to salt stress in three kinds of tissues. Transcript level of AhAQ1 accumulated rapidly within 1 h in the leaves and stems exposed to high salt stress, then decreased as the same level as the control 12 h and 24 h later, respectively. In the treated roots, AhAQ1 was transiently induced at 6 h after treatment (Figure 5). These results show that AhAQ1 is induced under salt stimulation, which suggests that it may be functional during the stress.
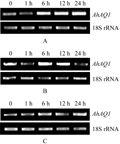 Figure 5 Tissue Expression pattern of AhAQ1 under salt stress Note: A~C: AhAQ1 expression in roots, stems and leaves after 0, 1 h, 6 h, 12 h, 24 h treatment with 250 mmol/L NaCl; 18S rRNA was used as the loading control of mRNA |
2 Discussion
Aquaporins contain six membrane-spanning domains linked by five loops and two conserved NPA motifs that lies in loop B and E, the mutation of which will lead to the loss of their function (Perston et al., 1994), Both their N- and C-terminal are located on the cytoplasmic side of the membrane. Since the amino acid sequence of AhAQ1 presented all the structure characters of aquaporin, it was normally considered to be an aquaporin in peanut.
Many studies conformed that water stress has a strong influence on aquaporin gene expression, and both induced, reduced, or unchanged patterns have been reported (Yamaguchi-Shinosaki et al., 1992; Baiges et al., 2002; Kawasaki et al., 2001; Grote et al., 1998 ). Three PIP genes, NtPIP1;1, NtPIP2;1 and NtAQP1 were isolated from tobacco plants. Under drought stress, the accumulation of NtPIP1;1 and NtPIP2;1 transcripts was significantly decreased, however that of the NtAQP1 transcript was increased (Mahdieh et al., 2008). The various expression patterns of aquaporins under water stress suggest that the molecular mechanism of plant tolerance to water stress is complex. AhAQ1 induced by salt stress in different tissues of peanut might confer the membrane permeability to water transport in water-deficient condition (Yamada et al., 1997).
Peanut is one of the most important oil crops in the world. Although many aquaporin genes were identified in different plants, but up to now there was no report on aquaporins in peanut. Severe salt stress leads to dramatic suppression of plant growth and development and causes a large loss of productivity during recent years. Our analysis suggests that the expression of AhAQ1 might function in plants’ response against salt stress. Further detailed investigations on this gene will provide comprehensive information of peanut salt stress tolerance and will show us a new way for molecular breeding leading to improve stress tolerance of agricultural crops.
3 Materials and methods
3.1 Plant materials and stress treatments
peanut seeds (Arachis hypogaea L. cv. Huayu28) were sown in sand and soil mixture (1:1), grown in a growth chamber with 16 h of light at 26°C, followed by 8 h of darkness at 22°C, which were watered daily. The plants at 3–leaf-stage were removed from the soil carefully to avoid injury and placed into a container with 250 mmol/L NaCl for 24 h. The roots,stems and leaves of the treated seedlings were sampled separately and stored at −80 °C at once at different time intervals after treatment.
3.2 RNA isolation and first strand cDNA synthesis
Total RNAs were extracted from various peanut tissues by using Trizol reagent (Tiangen) according to the manufacturer's protocol. The RNAs was sequentially treated with DNase I (Takara) at 37°C for 15 min in order to remove the remaining genomic DNA. The first strand cDNA was synthesized with 2 μg of purified total RNAs using the RT-PCR system (Promega) following the manufacturer's protocol.
3.3 Cloning of AhAQ1
To amplify the coding region of AhAQ1, two primers were designed. sense primer: 5’-CACACTTACCCATCTCATCAC-3’, antisense primer: 5’-CAATAATCAAGCACTTGCATT-3’. Each reaction system contained 2.5 μL of 10×PCR buffer with MgCl2, 0.5 μL of 10 μmol/L each primer, 1.0 μL of 20 mmol/L dNTPs, 1 μL cDNA samples and 0.5 μL Ex-Taq polymerase (Takara), and 19 μl double distilled water. PCR was performed on a DNA amplification machine (Effendorf) for an initial denaturizing at 95°C for 5 min, 28 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were run on 1% agarose gel and purified with Gel Extraction Kit (Takara) according to the manufacturer’s protocol. The purified products were then cloned into the pMD18-T Easy vector (Takara) and sequenced (Sangon, Shanghai).
3.4 Sequence analysis
The sequence analysis was performed using BioXM version 2.6 program. Multiple Sequence alignment obtained using the ClustalX ver 1.8 were displayed by the GENEDOC. Molecular phylogenies were computed using MEGA version 3.1. The TM prediction calculated with the hydrophobic values was completed by the program TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html).
3.5 Semi-quantitative RT-PCR analysis
The gene special primers, A1: 5’-ACCCTCCTCTTCCTCTACATC-3’ and A2: 5’-CAAGCACTGAGCCACCATAT-3’ were used for amplifying a fragment of 261 bp. Each reaction system contained 2.5 μL of 10×PCR buffer with MgCl2, 0.5 μL of 10 μmol/L each primer, 1.0 μL of 20 mmol/L dNTPs, 2 μL cDNA samples and 0.5 μL Ex-Taq polymerase (Takara), and 18 μL double distilled water. The thermal cycle used was as follows: 95°C for 5 min; 28 cycles of 95°C for 20 s, 55°C for 20 s, 72°C for 30 s, and a final 72°C for 10 min. As an internal control, a 226 bp PCR fragment of peanut 18S rRNA gene was amplified using primers 18S-F: 5’-ATTCCTAGTAAGCGCGAGTCATCAG-3’ and 18S-R: 5’-CAATGATCCTTCCGCAGGTTCAC-3’ (Wan et al., 2005). The PCR conditions for amplifying 18S rRNA gene in semi-quantitative RT-PCR assay are the same as those for AhAQ1 gene, but the annealing temperature was changed into 60°C. Three replicate PCR amplifications were performed for each sample.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (2006AA10A114), the National Key Basic Research and Development Project of China (2007CB116212).
References
Baiges I., SchÅffner A.R., Affenzeller M.J., and Mas A., 2002, Plant aquaporins, Physiol. Plant, 115(2): 175-182 doi:10.1034/j.1399-3054.2002.1150201.x
Chaumont F., Barrieu F., Wojcik E., Chrispeels M.J., and Jung R., 2001, Aquaporins constitute a large and highly divergent protein family in maize, Plant Physiol., 125(3): 1206-1215 doi:10.1104/pp.125.3.1206
Chaumont F., Moshelion M., and Daniels M.J., 2005, Regulation of plant aquaporin activity, Biol. Cell, 97(10): 749-764 doi:10.1042/BC20040133
Johanson U., and Gustavsson S., 2002, A new subfamily of major intrinsic proteins in plants, Mol. Biol. Evol., 19(4): 456-461
Johanson U., Karlsson M., Johansson I., Gustavsson S., SjÅvall S., Fraysse L., Weig A.R., and Kjellbom P., 2001, The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants, Plant Physiol., 126: 1358-1369 doi:10.1104/pp.126.4.1358
Katsuhara M., Koshio K., Shibasaka M., Hayashi Y., Hayakawa T., and Kasamo K., 2003, Over-expression of a barley aquaporin increased the shoot/root ratio and raised salt sensitivity in transgenic rice plants, Plant and Cell Physiology, 44(12): 1378-1383 doi:10.1093/pcp/pcg167
Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K., Galbraith D., and Bohnert H.J., 2001, Gene expression profiles during the initial phase of salt stress in rice, The Plant Cell, 13(4): 889-905
Lian H.L., Yu X., Ye Q., Ding X.S., Kitagawa Y., Kwak S.S., Su W.A., and Tang Z.C., 2004, The role of aquaporin RWC3 in drought avoidance in rice, Plant Cell Physiol., 45(4): 481-489 doi:10.1093/pcp/pch058
Luu D.T., and Maurel C., 2005, Aquaporins in a challenging environment: molecular gears for adjusting plant water status, Plant, Cell and Environment, 28(1): 85-96 doi:10.1111/j.1365-3040.2004.01295.x
Mahdieh M., Mostajeran A., Horie T., and Katsuhara M., 2008, Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants, Plant Cell Physiol., 49(4): 801-813 doi:10.1093/pcp/pcn054
Maurel C., Reizer J., Schroeder J.I., and Chrispeels M.J., 1993, The vacuolar membrane protein gamma-TIP creates water-specific channels in Xenopus oocytes, EMBO J., 12(6): 2241-2247
Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J.B., Engel A., and Fujiyoshi Y., 2000, Structural determinants of water permeation through aquaporin-1, Nature, 407(6804): 599-605 doi:10.1038/35036519
Preston G.M., Carroll T.P., Guggino W.B., and Agre P., 1992, Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein, Science, 256(5055): 385-387 doi:10.1126/science.256.5055.385
Sakurai J., Ishikawa F., Yamaguchi T., Uemura M., and Maeshima M., 2005, Identification of 33 rice aquaporin genes and analysis of their expression and function, Plant Cell Physiol., 46(9): 1568-1577 doi:10.1093/pcp/pci172
Siefritz F., Tyree M.T., Lovisolo C., Schubert A., and Kaldenho R., 2002, PIP1 plasma membrane aquaporins in tobacco: from cellular effects to function in plants, The Plant Cell, 14: 869-876 doi:10.1105/tpc.000901
van Hoek A.N., and Verkman A.S., 1992, Functional reconstitution of the isolated erythrocyte water channel CHIP28, J. Biol. Chem., 267(26): 18267-18269
Wan X., and Li L., 2005, Molecular cloning and characterization of a dehydration-inducible cDNA encoding a putative 9-cis-epoxycarotenoid dioxygenase in Arachis hygogaea L., DNA Seq., 16(3): 217-223 doi:10.1080/10425170500129785
Wang W., Vinocur B., and Altman A., 2003, Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance, Planta, 218(1): 1-14 doi:10.1007/s00425-003-1105-5
Yamada S., Komori T., Myers P.N., Kuwata S., Kubo T., and Imaseki H., 1997, Expression of plasma membrane water channel genes under water stress in Nicotiana excelsior, Plant Cell Physiol., 38(11): 1226-1231
Yamaguchi-Shinosaki K., Koizumi M., Urao S., and Shinozaki K., 1992, Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein, Plant Cell Physiol., 33(3): 217-224
Yu Q.J., Hu Y.L., Li J.F., Wu Q., and Lin Z.P., 2005, Sense and antisense expression of plasma membrane aquaporin from Brassica napus in tobacco and its effects on plant drought resistance, Plant Science, 169(4): 647-656 doi:10.1016/j.plantsci.2005.04.013
Zhu J.K., 2003, Regulation of ion homeostasis under salt stress, Curr. Opin. Plant Biol., 6(5): 441-445 doi:10.1016/S1369-5266(03)00085-2
. PDF(431KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Lijuan Pan
. Qingli Yang
. Yan Jiang
. shanlin Yu
Related articles
. Peanut
. Aquaporin
. Gene cloning
. Expression analysis
. Salt stress
Tools
. Email to a friend
. Post a comment


