Genetic Diversity of Faba Bean Germplasms in Qinghai and Core Germplasm Identified based on AFLP Analysis 
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2010, Vol. 1, No. 1 doi: 10.5376/lgg.2010.01.0001
Received: 21 May, 2010 Accepted: 11 Aug., 2010 Published: 13 Sep., 2010
Liu and Hou, 2010, Genetic Diversity of Faba Bean Germplasms in Qinghai and Core Germplasm Identified based on AFLP Analysis, Legume Genomics and Genetics Vol.1 No.1 (doi:10.5376/lgg.2010.01.0001)
In this study we analyzed the genetic diversities of Qinghai’s faba bean (Vicia faba L.) germplasms based on AFLP techniques. The results showed that the number of effective alleles in landraces, foreign germplasms and released cultivar are 1.065 5±0.126 3, 1.063 4±0.143 0, and 1.065 9±0.165 1, while the Shannon’s information index are 0.095 4±0.144 9, 0.084 3±0.151 8, 0.074 2±0.167 9, respectively. The ratio of effective allele of landrace is the lowest (73.73%), while its ploy-ratio is the highest (44.51%). Whereas the ratio of effective allele of released cultivars is the highest (90.39%), while its ploy-ratio is the lowest (17.92%). The genetic distance is relatively bigger in the landraces and smaller in the foreign germplasms. Genetic structure analysis revealed that the 149 faba bean germplasms in Qinghai can be divided into 7 groups. In this study we have selected 40 germplasms as core germplasm collection which would represent more than 80% genetic information of the tested 149 germplsams based on the genetic diversity and Nei\'s gene diversity.It is obvious that the core germplasm collection germplasms might be used as parent materials for hybrid breeding in faba bean breeding program.
AFLP (Amplified Fragment Length Polymorphism) is a new approach for detecting DNA polymorphism that was developed by Dutch scientists Zabeau and Vos in 1992. The AFLP procedures are generally as follows: first the total genomic DNA was digested and then was amplified by selective PCR. The specific joints were designed to add on the ends of amplified DNA fragments. Polymerase chain reaction (PCR) was performed with specific primers that bound the sequences of the given joints and neighboring restriction enzyme sites. Finally the amplified restriction fragments were scored on agarose gel electrophoresis (Zhou, 2005). AFLP is considered as an integration technology which possesses both the reliability of RFLP and the convenience of RAPD.
Qinghai is high altitude plateau province located in the west of mainland in China, where has a typical plateau continental climate with obvious vertical distribution. Faba bean is the traditional crops in Qinghai which has a long history of cultivation and wider regional distribution. The sowing area of faba bean reached 28 230 hm2. reported by the annual statistic report in Qinghai in 2009. Faba bean is well distributed at the ranges of the altitude from sea above 1 500 m to 3 000 m. There are abundant faba bean landrace and germplasms formed due to the domestication and cultivation in the unique ecological environment. Genetic diversity is recognized as a base for the germplasm utilization and crop genetical improvement. By evaluating the genetic diversity of the major faba bean germplasms with universal applicability and specificity can predict the variation degree in the separation progenies derived from pure breeding line. The methods based on the phenotypic and molecular markers were increasing used for revealing the diversity of germplasms of beans in the past decade, such as morphological marker used for analyzing the genetic diversity of faba germplsms (Wang et al., 2009; Zhang and Liu, 2007; Liu and Zong, 2008); AFLP markers were used to study the genetic diversity among 79 of inbred lines of faba bean which generated 477 polymorphic fragments (Zeid et al., 2001). Twentyeight accessions of faba bean inbred lines were employed to study the bean size diversity among the germplasm from European and Mediterranean by RAPD technique (Link et al., 1996). The variation of twenty-six faba bean populations from five geographic areas and five varietal types have been revealed by seven isozymes that the results supported the hypothesis of an isolating origination of the crop in Ethiopia and in Asian (Serradilla et al., 1993). Three isozymes, glutamate oxaloacetate transaminase (GOT), superoxide dismutase (SOD), and malic enzyme (ME), had been analysed electrophoretically to study the diversity within and between 33 faba bean (V. faba L. type minor) accessions, which were grouped as five groups with significant differences among them based on the parameters of principal components, hierarchical cluster, and canonical discri-minant analyses (Polignano et al., 1998). Core collection of broad bean in Qinghai was preliminary constructed based on morphological traits as well (Yang et al., 2009).
In this research we analyzed the genetic diversities of faba bean germplasms collected in Qinghai using AFLP method and constructed core faba germplasm collection based the data of genetic diversity. It might provide new insight for understanding the genetic basis of Qinghai faba bean germplasm as well as for better using the germplasm in breeding program.
1 Results
1.1 The polymorphism of AFLP primer analyzed
Ten pairs of primer combination can generated polymorphic bands in the ranges from 3 to 10 bands respectively, the primers combination named as E1/M6, E2/M2, E4/M4 and E7/M7 generated ten polymorphic bands and their polymorphic fragment ratio reached 52.63%, 52.63%, 50.00% and 52.63%, respectively, accordingly the proportion of effective alleles to alleles were respectively 67.80%, 68.46%, 73.35% and 68.59%. The pair of primer of E6/M6 produced three polymorphic bands only; Seven to eight polymorphic bands could be amplified from the rest primers combination (table 1).
|
Table 1 The polymorphic index for tested the AFLP primer combinations |
1.2 The AFLP polymorphism of faba bean collected in Qinghai
The average effective alleles were about 1.064 7 among 149 tested faba bean collections detected by using ten pairs of primers, which the proportion of effective alleles to alleles was 72.52%, as well as the polymorphic fragment ratio was 46.82% based on the total 81 amplified polymorphic bands, further Shannon’s information index was 0.094 6. The proportions of effective alleles to alleles were detected as 90.39%, 81.76% and 72.52% for released cultivars, introduced accessions and foreign germplasms, respectively. The polymorphism of local varieties were much higher with the 44.51% polymorph-ic fragment radio and 0.051 2 Nei’s gene diversity, whereas the one of released varieties was quite lower with the polymorphic fragment ratio and Nei’s gene diversity, which was 17.92% and 0.045 5 (table 2).
|
Table 2 The polymorphic index for tested Qinghai faba bean revealed by the AFLP primer combinations |
1.3 Genetic structure grouping analysis
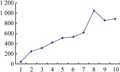
Genetic Structure of tested faba bean germplasms were analyzed by the software structure 2.2. The figure of K point of inflection was drawn based on the results of analysis (figure 1). There was the first obvious inflexion when K value was up to seven, which imply that the optimum grouping of tested faba bean in this research should be divided into seven groups (figure 2).
|
|
|
|
In this research 149 accessions were divided into seven groups, whereas there are different sources of germplasms scattered in different groups, which implied that there was distinct genetic variations and certain genetic similarities existed in the collection. The group one contains 26 populations, of which 50% released cultiv-ars, 16.67% foreign varieties and 12.87% landraces. The highest proportion of effective alleles to alleles was 92.96%, Shannon’s information index was 0.027 8, and the average genetic distance was very smaller among populations. The group two contains 24 populations, of which 18.81% landraces and being 13.89% foreign varieties. The proportion of effective alleles to alleles was 89.58%, and Shannon’s information index was 0.065 9. The third group three contains 20 popul-ations, of which 16.83% landraces, 5.56% foreign var-ieties and 8.33% released cultivars. The proportion of effective alleles to alleles was 84.47%, and Shannon’s information index was 0.124 0, and the largest genetic diversity existed among populations. The group four contains 22 populations, of which 14.85% landraces, 16.67% foreign varieties and 8.33% released cultivars. The proportion of effective alleles to alleles was 92.50%, and Shannon’s information index was 0.033 2, The group five contains 17 groups, of which 8.91% landraces, 16.67% foreign varieties and 16.67% released cultivars. The proportion of effective alleles to alleles was 92.50%, and Shannon’s information index was 0.064 5; The group six contains 18 groups, of which 8.91% landraces, 19.44% foreign varieties and 16.67% released cultivars. The proportion of effective alleles to alleles was 91.39%, and Shannon’s information index was 0.077 2; The group seven cont-ains 23 groups, of which 17.82% landraces and 11.11% foreign varieties. The proportion of effective alleles to alleles was 92.74%, and Shannon’s information index was 0.0 598 (table 3).
|
|
1.4 The genetic similarity of tested germplasms of faba bean
The genetic similarity coefficient of landrance, foreign varieties and released cultivars were calculated among 149 accessions, which were as 0.948 2 (0.867 1~1.000 0), 0.951 3 (0.884 4~1.000 0), 0.950 3 (0.907 5~0.994 2) respectively; whereas the genetic distance were as 0.053 5 (0~0.142 7)ã€Â0.050 3 (0~0.122 9), and 0.051 1 (0.005 8~0.097 0), respectively (figure 3). There is relatively higher genetic distance among landrances, while lower genetic distance among foreign varieties, which might be due to size and quantity of population.
|
|
1.5 Core collection of bean in Qinghai constructed based on AFLP
The phylogenetic tree of faba bean collection was established based on the germplasm origins with the aid of software Popgen V1.32 and MEGA 3.0. The core collection were assembled based on the phylogenetic tree and the targeted germplsams were proportional chosen with closer genetic relationship (table 4; figure 4). Total 40 accessions among 149 accessions of faba bean were selected as core collection germplasm that accordance rates at least reached 80% above in the number of alleles, effective alleles, polymorphisms bands, and coefficient of genetic similarity, genetic distance and gene diversity. Significance test results showed that 40 core collection germplasms of faba bean should be as representative in Qinghai which could cover whole genetic information due to no significant differences. Gene diversity (GD) and Nei’s gene diversity for the total faba bean collection were 0.058 3 and 0.103 2, respectively, which respectively increased 89.3% and 104.0%. The core collections contained 20 landraces, 11 foreign varieties and 9 released cultivars.
|
|
|
|
2 Discussion
2.1 Diversity analysis and germplasm classification
Germplasm resources of faba bean in Qinghai were abundant in molecular diversity, particular in faba landraces. The results are in agreement with those analyses of genetic diversity based on morphological traits using the same germplasm collections (Liu et al., 2009). But there was obvious difference from the result of germplasm classification. Using the Structure2.2 software, the genetic structure of faba bean in Qinghai was analyzed to determine that 149 accessions could be divided into seven groups, whereas three groups were clustered based on the phenotyping data. The classification results of germplasm resources were not only related to the types of markers but also were closely linked to classification method or foundation. The results indicated that classification of germplasm resources should not be completely classified by their origin. The same types of germplasm resources also had the genetic differences and certain genetic similarity.
The genetic distances among foreign germplasm were relatively small compared to other germplasm resources of faba bean in Qinghai. Therefore we suggested that introducing elite foreign germplasm to enrich genetic foundation of breeding of faba bean in Qinghai should be quite necessary.
2.2 Construction of core collection of faba bean in Qinghai
A core germplasm collection including 40 accessions was constructed from 149 accessions based on AFLP which contained genetic information accounting for over 80%. whereas a previous core germplasm collection including 21 accessions was constructed from 153 accessions of faba bean in Qinghai based on morphological traits by Yang et al. There are significant coincidences in the major characters between core collection and the whole tested samples, that was more than 90% (Yang et al., 2009). Although 149 accessions were chosen as test material from 159 of faba bean which were used by Yang et al., two core collections constructed by different markers were not the same at all. The same material and method were adopted by both sides, but their results were different. germplasms with special characters were fully considered in the morphologyical markers and while germplasm sources were considered in the AFLP maker. Core collections were constructed respectively based on released cultivars, foreign varieties and landraces. Therefore, in order to avoid the events of gene flow happen, it might be a smart way to use core collection in the genetic improvement, and also it is necessary to combine multiple methods to construct core collection.
3 Materials and methods
3.1 Materials
101 local landraces were used as research materials which collected from the different counties with greatly different ecological conditions (Haidong area, Haibei county, Hainan county and Huangnan county of Qinghai province). For comparison, 36 foreign varieties and 12 released cultivars were employed as reference research materials. Primer sequences were synthesized by Shanghai Biological Chemical Company (table 5).
|
|
3.2 Procedures of DNA extraction and AFLP Reac-tion protocols
The little modified CTAB procedure was applied to extract genomic DNA from faba bean (Liu et al., 2007b), and the AFLP Reaction protocols established by Liu etc was adopted in this research (Liu et al., 2007a).
3.3 Data Statistics and Analyses
Bands were scored by using 0 or 1, which 1 represented existing bands and 0 meant no band appeared. We used mixed model and relational model for occurrence frequency of allelic variation to analyze population genetic structure with the aids of software Structure 2.2. The given parameters of Structure (Burnin, Period and after Burnin) were fixed 10 000 times, and K value was set from 1 to 10. Each K value ran 10 times. The mean values of Var[lnP(D)] corresponded to each k value and [LnP(D)] were calculated, and then drew broken line figure to choose the best k value that the numbers of population genetic structure could be yielded (Liu et al., 2009). The effective allelic variation, the polymorphic fragment percentage, the diversity index of Shannon weaver and genetic distance were calculated by Popgen1.32 software; Core collection was structured by using clustering method (Yang et al., 2009; Wang et al., 2002).
Authors’ contributions
Yujiao Liu designed and implemented the experiment, and also drafted the manuscript. Wanwei Hou participated in partial experimental work. Both authors read and approved the final manuscript.
Acknowledgements
This research was jointly supported by National Key Technology R&D Program (2007BAD59B05), Special Fund of The Industrial Technology System Construction of Modem Agriculture and Countermeasures (nycytx18-G2), and Key Technologies R & D Program of Qinghai Province (2006-G-126).
References
Gemechu K., Mussa J., and Tezera W., 2005, Extent and pattern of genetic diversity for morpho-agronomic traits in ethiopian highland pulse landraces II, faba bean (Vicia faba L.), Genetic Resources and Crop Evolution, 52(5): 551-561 doi:10.1007/s10722-003-6022-8
Liu J., Guan J.P., Xu D.X., Zhang X.Y., Gu J., and Zong X.X., 2008, Phenotypic diversity of Lentil (Lens culinaris Medik.) germplasm resources, Journal of Plant Genetic Resources, 9(2): 173-179
Liu X.J., Ren J.Y., Zong X.X., Guan J.P., and Zhang X.Y., 2007, Establishment and optimization of AFLP for faba bean, Journal of Plant Genetic Resources, 8(2): 153-158
Liu Y.J., and Zong X.X., 2008, Morphological diversity analysis of faba bean (Vicia Faba L.), Germplasm resources from Qinghai, Journal of Plant Genetic Resources, 9(2): 79-83
Polignano G.B., Quintano G., and Bisignano V., 1998, Enzyme polymorphism in faba bean (Vicia faba L. minor) accessions, genetic interpretation and value for classification, Euphytica, 102(2): 169-176 doi:10.1023/A:1018340228500
Polignano G.B., Uggenti P., and Scippa G., 1993, The pattern of genetic diversity in faba bean collections from Ethiopia and Afghanistan, Genetic Resources and Crop Evolution, 40(2): 71-75 doi:10.1007/BF00052637
Serradilla M.T., De Mora M., and Moreno T., 1993, Geographic dispersion and varietal diversity in Vicia faba L., Genetic Resources and Crop Evolution, 40(3): 143-151 doi:10.1007/BF00051119
Wang S.M., Cao Y.S., and Hu J.P., 2002, Preliminary establishment of Chinese Adzuki bean germplasm resource core collection with agroecology and characterization data, Acta Agriculturae Boreali-Sinica, 17(1): 35-40
Wang X.J., Qi X.S., and Wang X.R., 2009, Genetic diversity analysis of main agronomic characters in faba bean germplasm, Research of Agricultural Modernization, 30(4): 633-636
Yang J., Chi D.Z., and Liu Y.J., 2009, Preliminary construction of core collection of broad bean in Qinghai based on morphological traits, Molecular Plant Breeding, 7(3): 599-606
Zeid M., Schön C.C., and Link W., 2001, Genetic diversity in a group of recent elite faba bean lines, Czech J. Gene. Plant Breed., (37): 34-40
Zhang X.T., and Liu Y.J., 2009, Morphological diversity analysis of faba bean [Vicia faba L.] germplasm resources, Qinghai Science Technology, 5: 17-19
Zhou Y.Q., 2005, The application of DNA marker for researched plant, Chemical Industry Press, Beijing, China, pp.162-165
Zong X.X., Guan J.P., Wang S.M and Liu Q.C, 2008, Genetic diversity Chinese pea (Pisum sativum L.) landrace revealed by SSR markers, Acta Agronomic Sinica, 34(8): 1330-1338 doi:10.1016/S1875-2780(08)60045-0
Link W., Dixkens C., and Singh M., 1996, Genetic diversity in European and Mediterranean faba germplasm revealed by RAPD markers, Theor. Appl. Genet., (92): 637-643
Liu Y.J., Yang J., and Cui Z.S., 2007, Modified DNA extraction for AFLP analysis in faba bean, Molecular Plant Breeding, 5(5): 747-750
. PDF(285KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yujiao Liu
. Wanwei Hou
Related articles
. Faba bean ( Vicia faba L.)
. Germplasm
. Core germplasm
. AFLP
Tools
. Email to a friend
. Post a comment

.png)



