Isolation and Expression Analysis of a β-Ketoacyl-Acyl Carrier Protein Synthase â… gene from Arachis hypogaea L. 
 Author
Author  Correspondence author
Correspondence author
Legume Genomics and Genetics, 2010, Vol. 1, No. 3 doi: 10.5376/lgg.2010.01.0003
Received: 01 Jul., 2010 Accepted: 10 Sep., 2010 Published: 10 Oct., 2010
Chi et al., 2010, Isolation and Expression Analysis of a β-Ketoacyl-Acyl Carrier Protein Synthase â… gene from Arachis hypogaea L., Legume Genomics and Genetics Vol.1 No.3 (doi: 10.5376/lgg.2010.01.0003)
β-ketoacyl-acyl carrier protein synthase (KAS) plays a pivotal role in de novo fatty acid biosynthesis, joining short carbon units to construct fatty acyl chains by a three-step Claisen condensation reaction in plants and bacteria. A putative KASâ… cDNAs was isolated from peanut by searching a peanut seedling full-length cDNA library. The AhKASâ… contains four strictly conserved residues Cys221–His361–Lys392–His397 in the active site, which is an important characteristic of KAS Is in plants and bacteria. The AhKASâ… gene containing 1 912 bp cDNA sequence with a 1 413 bp ORF, encodes 470 amino acids. The predicted amino acid sequences translated from the AhKASâ… gene shared 90.2% sequence identity to the corresponding protein in Glycine max. The cDNA was cloned into plasmid pET-28a and expressed in Escherichia coli BL21. Quantitative real-time PCR analysis suggested AhKASâ… was expressed with higher levels in leaf and seed than those in other tissues. In addition, AhKASâ… RNA was found in high abundance at 45 and 65 DAP (days after pegging) during seed development. This work may serve as a foundation for further studies on the mechanisms regulating the expression of KASâ… gene and provide candidate genes for modifying oil quality via transgenic plants.
The de novo fatty acid biosynthesis is a very important primary metabolic pathway, producing palmitic acid (16:0) and stearic acid (18:0) that serve as the precursors for other fatty acids with different lengths and saturation levels (Ohkrigge and Browse, 1995). Fatty acid biosynthesis in higher plants is mainly catalyzed by a suit of enzymes in plastids and the two-carbon elongation reactions are catalyzed by β-ketoacyl-acyl carrier protein (ACP) synthase (KAS, EC 2.3.1.41) family. KASâ…¢ initiates the fatty acid synthesis to form 4:0-ACP in plants by catalyzing the condensing reaction of acetyl-CoA and malonyl-ACP. KASâ… catalyzes the condensations of acetate unites to a growing acyl-ACP leading to the synthesis of palmitoyl-ACP (16:0-ACP). KASâ…¡ is responsible for the elongation of 16:0-ACP to 18:0-ACP (Shimakata and Stumpf, 1982). A fourth KAS enzyme (KASâ…£) located in plastid specific for the synthesis of medium-chain acids has also been reported (Siggaard-Andersen et al., 1994; Dehesh et al., 1998). In addition, the mitochondrial condensing enzyme (mtKAS), which catalyzes all the condensation reactions in mitochondrial fatty acid synthesis, has been characterised from A. thaliana (Yasuno et al., 2004) and the crystal structure of this enzyme has been determined (Olsen et al., 2004).
By now, several species of β-ketoacyl-ACP synthases in plants and bacteria have been identiï¬ed, distinct in amino acid sequence, chain length speciï¬city for their substrates and sensitivity to cerulenin, an inhibitor of condensing enzymes (Vance et al., 1972;Kauppinen et al., 1988). The KAS family (fabB, fabF and fabH) have been most extensively studied in E. coli and their crystal structures have been determined (White et al., 2005). In contrast to the well-studied E. coli KAS family, plant KAS family is largely uncharacterized except in A. thaliana (Li et al., 2009). KASâ…¢ was first purified to homogeneity from spinach (Clough et al., 1992) and its cDNAs have been cloned from several plant species (Tai and Jaworski, 1993; Tai et al., 1994; Slabaugh et al., 1995; Chen and Post-Beittenmiller, 1996). In addition, cDNAs of KASâ…¡ have also been isolated from many species including soybean, rape, perilla and Arabidopsis (Aghoram et al., 2006; Carlsson et al., 2002; Hwang et al., 2000). It has been demonstrated that the altered expression levels of KASâ…¡ and KASâ…¢ lead to change of oil content and qualities in A. thaliana (Abbadi et al., 2000;Dehesh et al., 2001; Pidkowich et al., 2007). KASâ… has been purified from Spinacia oleracea leaves, and a gene encoding KASâ… has also been characterized in barley (Shimakat et al., 1983; Kauppinen, 1992). However, the function and the regulation of KAS family were still not well understood in higher plants.
Peanut is one of the four most important oil crops widely grown in the world. It would be of great importance to study the fatty acid biosynthesis pathway for improving oil quality and increasing oil content of peanut. In this study, we isolated and characterized a cDNA containing the complete coding region of KASâ… gene, and analyzed its expression in different organs and at different developmental stages of seeds.
1 Results
1.1 Molecular cloning of AhKASâ… gene from peanut
One full-length AhKASâ… cDNA clone was identiï¬ed from a peanut seedling full-length cDNA library (unpublished data) based on the amino acid similarity. The AhKASâ… gene is 1 912 bp in length containing a 1 413 bp ORF, starting with an initiating codon at 238 bp and ending with a stop codon at 1 650 bp (accession number FJ768729). The predicted protein product of AhKASâ… comprises 470 amino acids with the calculated molecular mass of 49.958 9 kD and a pI of 8.46. Prediction of subcellular location suggested that AhKASâ… protein probably located in chloroplast. The first 48 amino acids at the N-terminal end of the deduced protein have a high proportion of hydroxylated and small, hydrophobic amino acids, typical of chloroplast transit peptide. We have tentatively identified the transit peptide cleavage site at amino acid 49 based on the ChloroP 1.1 Server. A Blast search revealed that the primary structure of AhKASâ… shared 90.2%, 84.8%, 84.5%, 81.3% identity with KASâ… genes from Glycine max, Ricinus communis, Helianthus annuus and Arabidopsis thaliana, respectively (Figure 1). In addition, using Rapid Amplification of cDNA Ends (RACE) method, we have isolated a KASâ…¡ gene (accession number FJ358425) containing a 1 521 bp complete open reading frame (ORF) from peanut seedling, named AhKASâ…¡, which was presumed to be located in chloroplast. The plastid-localized AhKASâ… and AhKASâ…¡ are 50.3% identical in this study, compared with 38% in E. coli. AhKASâ… shared 28.8% and 38.5% identity with EcFabB and EcFabF, respectively, whereas AhKASâ…¡ shared 26.7% and 35.8% identity.
1.2 Sequence and phylogenetic analysis of AhKASâ… gene
A subgroup of β-ketoacyl-ACP synthases, including mitochondrial β-ketoacyl-ACP synthase, bacterial plus plastid β-ketoacyl-ACP synthases â… and â…¡, and a domain of human fatty acid synthase, have a Cys-His-His triad and also a completely conserved Lys in the active site, which are referred to as the CHH group (von Wettstein-Knowles et al., 2006). Another group of decarboxylating condensing enzymes called CHN (N for asparagine), represented by KASâ…¢ and certain polyketide synthases (Qiu et al., 1999; Davies et al., 2000; Scarsdale et al., 2001) with an active site composed of a cysteine nucleophile, a histidine and an asparagine. Four conserved residues Cys221–His361–Lys392 –His397 of AhKASâ… were revealed by sequence alignments (Figure 1). The CHH active site (Cys–His–His) was highly conserved in KASâ… and KASâ…¡ in higher plants and E. coli (Li et al., 2009).
|
Figure 1 Alignment of the complete deduced amino acid sequences of β-ketoacyl-ACP synthase â… genes
|
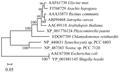
To examine the relationships among different sources of KASâ… genes, the neighbour-joining method was used to construct the phylogenetic trees (Figure 2). All tree topologies are highly congruent. As shown in the phylogenetic tree, all of the AhKASâ… genes fell into two subfamilies: the bacteria subfamily and the cyanobacteria/green algae/mosses/higher plants subfamily. The AhKASâ… gene from peanut clustered with those from higher plants, and the genes from cyanobacteria may be the origin of genes from higher plants, mosses and eukaryotic algae.
|
Figure 2 Neighbor-joining tree based on the deduced amino acid sequences of KASâ… homologs
|
1.3 Heterologous expression of AhKASâ… gene in E. coli cells
The AhKASâ… enzyme was overexpressed using the pET vector system in E. coli. Gene expression and the molecular weight of this enzyme were monitored by SDS-PAGE (Figure 3). After IPTG induction, a novel polypeptide with the molecular mass of approximately 49 kD was expressed in E. coli as revealed by SDS-PAGE. The recombinent protein reached maximal expression levels 4 to 8 h after induction and showed a similar molecular weight of approximately 49 kD compared to the deduced value.
|
Figure 3 The SDS-PAGE map of AhKASâ… protein expression
|
1.4 Quantitative real-time PCR analysis of AhKASâ… gene
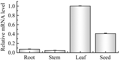
The quantitative real-time PCR (qRT-PCR) was employed to confirm the expression patterns of AhKASâ… gene in four peanut tissues and at different developmental stages of seeds. β-actin was used as an internal reference control for total RNA input. β-actin PCR product was not detected when reverse transcriptase was omitted, indicating that the RNA template was free of genomic DNA. The results revealed that AhKASâ… gene displayed tissue-specific expression patterns in peanut (Figure 4). The AhKASâ… gene was expressed dominantly in leaf among four tissues tested, and expressed at the lowest level in stem. In addition, comparative analysis among seeds over five development stages showed that AhKASâ… gene expressed in an irregular course during seed development. It had highest expression at 45 DAP and the expression level at 65 DAP was also relatively high (Figure 5).
|
Figure 4 Expression analysis of AhKASâ… gene in four different tissues
|
|
|
2 Discussion
The formation of carbon–carbon bonds is a fundamental biochemical reaction. A number of enzymes involved in various biosynthetic pathways accomplish this reaction by different means. Among them, one mechanism is the Claisen condensation, a reaction catalyzed by β-ketoacyl-ACP synthase (KAS) enzymes (Olsen et al., 2001). In higher plants ï¬ve types of KAS, namely, KASâ… , KASâ…¡, KASâ…¢, KASâ…£ and mitochondrial KAS, have been reported (Li et al., 2009). In the present study, we isolated a KASâ… orthologue in peanut seedling. We investigated the homology of this gene, the deduced protein’s active sites, physicochemical properties and its subcellular location through bioinformatics analysis. The results revealed that the amino acid sequences of peanut AhKASâ… shared high sequence identity, 90.2% and 84.8%, with Glycine max and Ricinus communis KASâ… proteins, respectively. Moreover, phylogenetic analysis showed that AhKASâ… gene clustered with those from higher plants, and the genes from cyanobacteria may be the origin of genes from higher plants, mosses and eukaryotic algae. Real-time PCR analysis revealed that the expression level of AhKASâ… was higher in leaf and seed than those in other tissues. In addition, AhKASâ… RNA was found in high abundance at 45 and 65 DAP during seed development.
Plant seed oil production can be manipulated through modifying the plant type â…¡ FAS components. Despite recent progress in detailed characterization of many enzymes involved in plant fatty acid synthesis, the mechanism of plant fatty acid synthesis is not well understood (Ohlrogge and Jaworski, 1997). To increase the expression of a single gene in a complex fatty acid synthesis pathway is not effective in changing the final product, unless concomitant changes in other enzymes involved are achieved. Therefore the regulation should be spread out and coordinated among the many enzymes involved in the pathway (Dehesh et al., 2001). In the present study, the β-ketoacyl-ACP synthase â… gene from peanut was identified. This enzyme is critical to the elongation step and plays a pivotal role in the regulation of the entire pathway (Magnuson et al., 1993). This work may serve as a foundation for further studies on the mechanisms regulating the expression of KASâ… gene and provide candidate genes for modifying oil quality via transgenic plants.
3 Materials and methods
3.1 Plant materials
Peanut seeds (Arachis hypogaea L. cultivar Huayu19) were sown in sand and soil mixture (1:1), grown in a growth chamber under a 16 h-8 h light-dark cycle at 26°C and 22°C, respectively. Three kinds of 12-day-old tissues including root, stem and leaves were collected as experimental materials for quantitative real-time RT-PCR analysis. In addition, the immature peanut seeds from 25 to 60 days after pegging (DAP) were also collected for expression analysis. The peanut cultivar was provided by Shandong Peanut Research Institute, Qingdao, China.
3.2 Nucleic acid manipulation
Total RNA was extracted from samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA samples were used for real-time RT-PCR after RQ1 RNase-free DNaseI (Promega, Wisconsin, USA) treatment to remove genomic DNA. The ï¬rst-strand cDNA was synthesized with RT-PCR kit (Promega, Wisconsin, USA) using 500 ng of total RNA according to the manufacturer’s instructions. Controls received water instead of reverse transcriptase to assess any contamination from genomic DNA as described by Zhou et al. (2007).
3.3 Full-length cDNA sequence isolation
PCR was performed with the LA PCR system (TaKaRa) using 2.5 μL of 10×PCR buffer with MgCl2, 1 μL of 10 μmol/L each primer, 4.0 μL of 10 mmol/L dNTPs, 1 μL cDNA samples and 0.5 μL LA Taq™ DNA polymerase, and 15 μL double distilled water. Reaction conditions were as follows: 5 min denaturation at 94°C, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min 30 s, ï¬nally extended at 72°C for 10 min. The PCR products were run on 1% agarose gel and purified with Gel Extraction Kit (TaKaRa) according to the manufacturer’s protocol. The purified products were then cloned into the pMD18-T Easy vector (TaKaRa) and sequenced (Shangon, Shanghai).
3.4 Sequence analysis
Open reading frame (ORF) and encoded amino acid sequence of genes were deduced by BioXM 2.6. Physicochemical properties of the deduced protein were predicted by Protparam (http://www.expasy.ch
/tools/protparam.html). The putative subcellular localizations of the candidate proteins were estimated by TargetP (http://www.cbs.dtu.dk/services/TargetP/) and Predotar (http://urgi.versailles.inra.fr/predotar/
predotar.html). The potential N-terminal presequence cleavage site was predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
3.5 Phylogenetic analysis
Amino acid sequences were aligned using ClustalX program with the implanted BioEdit (Thompson et al., 1994). The neighbor-joining (NJ) method in MEGA4 (Tamura et al., 2007) was used to construct the phylogenetic tree. Bootstrap with 1 000 replicates was used to establish the confidence limit of the tree branches. Default program parameters were used.
3.6 Expression of AhKASâ… in E. coli
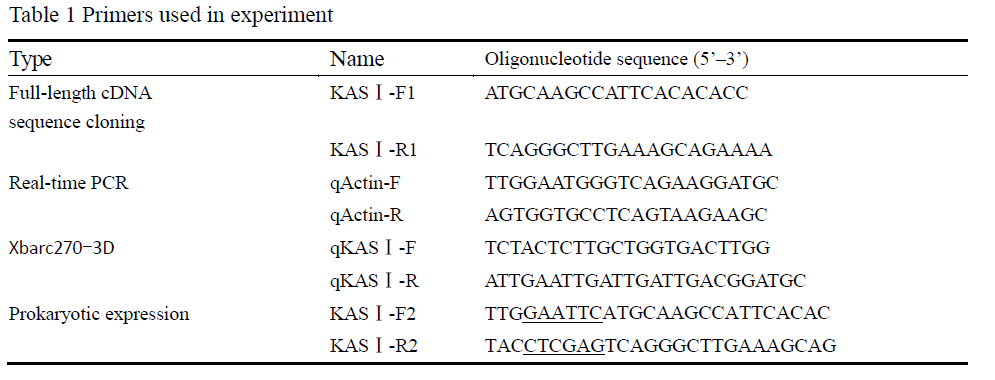
Two speciï¬c primers (KASâ… -F2 and KASâ… -R) were used to obtain the full-length open reading frames (Table 1). The 5’ end of the KASâ… -F2 and KASâ… -R2 contains an EcoRâ… or an Xhoâ… restriction site (underlined) to facilitate subsequent manipulations. The ampliï¬ed fragment was digested with EcoRâ… and Xhoâ… , followed by ligation with EcoRâ… /Xhoâ… -digested pET-28a (Novagen) to produce pET-KASâ… , and no amplification errors were detected through re-sequencing. The constructed plasmid was transformed into E. coli strain BL21 and grown in 50 mL LB medium containing 50 μg/mL kanamycin at 37°C to a optical density of A600=0.6~0.8. The expression of recombinant protein was induced by adding isopropyl β-D-thiogalactoside (IPTG) to the culture at a ï¬nal concentration of 1.0 mmol/L. Cells were harvested by centrifugation after incubation at 37℃ for 1 h, 2 h, 4 h, 6 h, 8 h, respectively. The protein extracts were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie brilliant blue R-250.
3.7 Quantitative real-time PCR
The real-time PCR analysis was performed by using a LightCycler 2.0 instrument system (Roche, Germany). β-actin gene was taken as reference gene. Three pairs of gene-speciï¬c primers (Table 1) were designed according to the AhKASâ… cDNA (qKASâ… -F and qKASâ… -R) and β-actin (qActin-F and qActin-R) sequences. The real-time RT-PCR reactions were performed by using the SYBR Premix Ex Taq polymerase (TaKaRa, Japan) according to the manufacturer’s instructions. The expression of the gene was calculated relative to the calibration sample and the β-actin to normalize the sample input amount. All the experiments were performed in triplicate to ensure the data accuracy.
|
|
Authors’contributions
Xiaoyuan Chi was responsible for the large part of data acquisition. Mingna Chen, Qingli Yang, Ya’nan He, Lijuan Pan, Yuan Gao analyzed data and wrote substantial parts of the paper. Shanlin Yu conceived the overall study, performed the experiment designs and took part in the data analysis and the writing. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by a grant from Modern Agro-industry Technology Research System (nycytx-19), National High-Tech Research and Development Plan of China (2006AA10A114; 2007AA10Z189), National Project of Scientific and Technical Supporting Program (2008BAD97B04), the National Natural Science Foundation of China (31000728) and the Natural Science Fund of Shangdong Province (ZR2009DQ004)
References
Abbadi A., Brummel M., and Spener F., 2000, Knockout of the regulatory site of 3-ketoacyl-ACP synthase IIIenhances short- and medium-chain acyl-ACP synthesis, Plant J., 24: 1-9 doi:10.1046/j.1365-313x.2000.00841.x
Aghoram K., Wilson R.F., Burton J.W., and Dewey R.E., 2006, A mutation in a 3-keto-acyl-ACP synthase II gene is associated with elevated palmitic acid levels in soybean seeds, Crop Sci., 46: 2453-2459 doi:10.2135/cropsci2006.04.0218
Carlsson A.S., LaBrie S.T., Kinney A.J., von Wettstein-Knowles P., and Browse J., 2002, A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket, Plant J., 29: 761-770 doi:10.1046/j.1365-313X.2002.01253.x
Chen J., and Post-Beittenmiller D., 1996, Molecular cloning of a cDNA encoding β-ketoacyl-acyl carrier protein synthase III from leek, Gene, 182(1-2): 45-52
Clough R.C., Matthis A.L., Barnum S.R., and Jaworski J.G., 1992, Purification and characterization of 3-ketoacyl-acyl carrier protein synthase-III from spinach: a condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis, J. Biol. Chem., 267: 20992-20998
Davies C., Heath R.J., White S.W., and Rock C.O., 2000, The 1.8 Å crystal structure and active site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli, Structure, 8: 85-195 doi:10.1016/S0969-2126(00)00094-0
Dehesh K., Edwards P., Fillatti J., Slabaugh M., and Byrne J., 1998, KASIV: a 3-ketoacyl-ACP synthase from Cuphea sp. is a medium chain specific condensing enzyme, Plant J., 15(3): 383-390 doi:10.1046/j.1365-313X.1998.00218.x
Dehesh K., Tai H., Edwards P., Byrne J., and Jaworski J.G., 2001, Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis, Plant Physiol., 125: 1103-1114 doi:10.1104/pp.125.2.1103
Hwang S.K., Kim K.H., and Hwang Y.S., 2000, Molecular cloning and expression analysis of 3-ketoacyl-ACP synthases in the immature seeds of Perilla frutescens, Mol. cells, 10(5): 533-539 doi:10.1007/s10059-000-0533-3
Kauppinen S., Siggaard-Andersen M., and von Wettstein-Knowles P., 1988, β-Ketoacyl-ACP synthase I of Escherichia coli: nucleotide sequence of the fabB gene and identiï¬cation of the cerulenin binding residue, Carlsberg Res., Commun., 53: 357-370 doi:10.1007/BF02983311
Kauppinen S., 1992, Structure and expression of the Kas12 gene encoding a β-ketoacyl-acyl carrier protein synthase I isozyme from barley, J. Biol. Chem., 267(25): 23999-24006
Li M.J., Li A.Q., Xia H., Zhao C.J., Li C.S., Wan S.B., Bi Y.P., and Wang X.J., 2009, Cloning and sequence analysis of putative type II fatty acid synthase genes from Arachis hypogaea L., J. Biosci., 34(2): 227-238 doi:10.1007/s12038-009-0027-1
Magnuson K., Jackowski S., Rock C.O., and Cronan J.E.Jr., 1993, Regulation of fatty acid biosynthesis in Escherichia coli, Microbiol. Rev., 57: 522-542
Ohkrigge J., and Browse J., 1995, Lipid biosynthesis, The Plant Cell, 7: 957-970
Ohlrogge J.B., and Jaworski J.G., 1997, Regulation of fatty acid synthesis, Annu. Rev. Plant Physiol. Plant Mol. Biol., 48:109-136 doi:10.1146/annurev.arplant.48.1.109
Olsen J.G., Kadziola A., von Wettstein-Knowles P., Siggaard-Andersen M., and Larsen S., 2001, Structures of β-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery, Structure, 9(3): 233-243 doi:10.1016/S0969-2126(01)00583-4
Olsen J.G., Rasmussen A.V., von Wettstein-Knowles P., and Henriksen A., 2004, Structure of the mitochondrial beta-ketoacyl-[acyl carrier protein] synthase from Arabidopsis and its role in fatty acid synthesis, FEBS Lett., 577: 170-174 doi:10.1016/j.febslet.2004.10.007
Pidkowich M.S., Nguyen H.T., Heilmann I., Ischebeck T., and Shanklin J., 2007, Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil, Proc. Natl. Acad. Sci., USA., 104(11): 4742-4747 doi:10.1073/pnas.0611141104
Qiu X., Janson C.A., Konstantinidis A.K., Nwagwu S., Silverman C., Smith W.W., Khandekar S., Lonsdale J., and Abdel-Meguid S.S., 1999, Crystal structure of β-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis, J. Biol. Chem., 274(51): 36465-36471 doi:10.1074/jbc.274.51.36465
Scarsdale J.N., Kazanina G., He X., Reynolds K.A., and Wright H.T., 2001, Crystal structure of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthase III, J. Biol. Chem., 276: 20516-20522 doi:10.1074/jbc.M010762200
Shimakata T., and Stumpf P.K., 1982, Isolation and function of spinach leaf beta-ketoacyl-[acyl-carrier-protein] synthases, Proc. Natl. Acad. Sci., USA., 79(19): 5808-5812 doi:10.1073/pnas.79.19.5808
Siggaard-Andersen M., Wissenbach M., Chuck J.A., Svendsen I., Olsen J.G., and von Wettstein-Knowles P., 1994, The fabJ-encoded β-ketoacyl-[acylcarrier protein] synthase IV from Escherichia coli is sensitive to cerulenin and specific for short-chain substrates, Proc. Natl. Acad. Sci., USA., 91: 11027-11031 doi:10.1073/pnas.91.23.11027
Slabaugh M.B., Tai H., Jaworski J., and Knapp S.J., 1995, cDNA clones encoding beta-ketoacyl-acyl carrier protein synthase III from Cuphea wrightii, Plant Physiol., 108: 443-444 doi:10.1104/pp.108.1.443
Tai H.Y., and Jaworski J.G., 1993, 3-Ketoacyl-acyl carrier protein synthase III from spinach (Spinacia oleracea) is not similar to other condensing enzymes of fatty acid synthase, Plant Physiol., 103: 1361-1367 doi:10.1104/pp.103.4.1361
Tai H., Post-Beittenmiller D., and Jaworski J.G., 1994, Cloning of a cDNA encoding 3-ketoacyl-acyl carrier protein synthase III from Arabidopsis, Plant Physiol., 106(2): 801-802 doi:10.1104/pp.106.2.801
Tamura K., Dudley J., Nei M., and Kumar S., 2007, MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0, Molecular Biology and Evolution., 24: 1596-1599 doi:10.1093/molbev/msm092
Thompson J.D., Higgins D.G., and Gibson T.J., 1994, CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res., 22(22): 4673-4680 doi:10.1093/nar/22.22.4673
Vance D.E., Goldberg I., Mitsuhashi O., and Bloch K., 1972, Inhibition of fatty acid synthetases by the antibiotic cerulenin, Biochem. Biophys. Res. Commun., 48(3): 649-656 doi:10.1016/0006-291X(72)90397-X
von Wettstein-Knowles P., Olsen J.G., McGuire K.A., and Henriksen A., 2006, Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type β-ketoacyl-acyl carrier protein synthases, FEBS J., 273(4): 695-710 doi:10.1111/j.1742-4658.2005.05101.x
White S.W., Zheng J., Zhang Y.M., and Rock C.O., 2005, The structural biology of type II fatty acid biosynthesis, Annu. Rev. Biochem., 74: 791-831 doi:10.1146/annurev.biochem.74.082803.133524
Yasuno R., von Wettstein-Knowles P., and Wada H., 2004, Identiï¬cation and molecular characterization of the beta-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase, J. Biol. Chem., 279(9): 8242-8251 doi:10.1074/jbc.M308894200
Zhou X.R., Robert S.S., Petrie J.R., Frampton D.M., Mansour P.M., Blackburn S.I., Nichols P.D., Green A.G., and Singh S.P., 2007, Isolation and characterization of genes from the marine microalga Pavlova salina encoding three front-end desaturases involved in docosa- hexaenoic acid biosynthesis, Phytochemistry, 68: 785-796 doi:10.1016/j.phytochem.2006.12.016
. PDF(2545KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Xiaoyuan Chi
. Mingna Chen
. Qingli Yang
. Ya'nan He
. Lijuan Pan
. Yuan Gao
. Shanlin Yu
Related articles
. Fatty acid biosynthesis
. β-ketoacyl-ACP synthase â…
. Expression analysis
. Peanut ( Arachis hypogaea L.)
Tools
. Email to a friend
. Post a comment